Translate this page into:
Use of cost-effective software for lesion localization in brain surgery: Technical note
*Corresponding author: Deepak Kumar Jha, Department of Neurosurgery, All India Institute of Medical Sciences, Jodhpur, Rajasthan, India. drdeepakjha@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Jha K, Harsh M, Agrawal M, Saini L, Tiwari S, Jha DK. Use of cost-effective software for lesion localization in brain surgery: Technical note. J Neurosci Rural Pract. 2024;15:361-4. doi: 10.25259/JNRP_546_2023
Abstract
Lesion localization has been an important aspect of neurosurgery and has advanced significantly with technological evolution. The journey started from the localization of lesion based on clinical findings to the current era where neuronavigation and virtual reality are being used for the purpose. However, the financial implications of these advanced equipments have made them inaccessible for patients in the majority of low- and middle-income countries. The authors describe techniques to use software, which are cost effective and can be used effectively for the localization of a lesion of the brain.
Keywords
Software
Brain surgery
Neurosurgery
Lesion localization
INTRODUCTION
Lesion localization in neurosurgery is an important aspect of successful treatments. Clinical finding based neurosurgical localization of lesions has been replaced by gradually advancing technological advancements, which include imaging studies, a variety of stereotactic or localization frames, neuronavigation, robotics integrated with neuronavigation, and most recently augmented and virtual reality techniques.[1-7] Unfortunately, technological advancements have gradually made neurosurgical services costlier, and access for patients of low- and middle-income countries (LMICs) is a genuine concern mainly because of financial implications.
The authors describe a technique to use a software application on a laptop or desktop computer for easy localization of lesions in the brain during neurosurgery.
TECHNIQUE
A digital file of contrast-enhanced (CE) magnetic resonance imaging (MRI) of the brain in the Digital Imaging and Communications in Medicine (DICOM) format is used for the technique. Authors use RadiAnt DICOM viewer, version 2020.2.3 (Medixant, Poznań, Poland) on a laptop or desktop [Figure 1]. Detailed technique to use it is described in our earlier report.[8] Three-dimensional (3D) images can be seen using clicking the “3D” icon of the window of the RadiAnt DICOM viewer [Figure 1]. An MRI sequence is selected in which the image is seen most clearly and usually, it is an CE sequence if the lesion is contrast enhancing. A T2 3D fluid-attenuated inversion recovery (FLAIR) sequence usually shows the lesion even if it does not show on the CE sequence. It is important to get these two sequences in 3D form (continuous or zero gap). For localization purposes during surgery, 3D T2 FLAIR sequences are used whereas the CE sequence shows vessels (arteries and veins) over the brain surface and contrast-enhancing lesions.

- RadiAnt DICOM viewer window where “3D sagittal T2 fluid-attenuated inversion recovery” sequence is selected. Arrows show icons of “Adjust image window,” “3D volume rendering technique,” and “3D multiplanar reformation.”
Three dimensions of multiplanar reformation (MPR) (MPR icon on top of the window) [Figure 1] images are used to corroborate the image in the plane, which corresponds best to the brain surface with lesion or lesion underlying it [Figure 2a]. The extent of the lesion over the brain surface is identified by shifting the three axes. The extent of the lesion should be marked over the volume rendering technique (VRT) image of the brain surface, taking reference to the 3D MPR images [Figures 2b and c]. The T2 3D FLAIR sequence VRT images reveal brain surface sulci and gyri and the lesion also by changing the window setting [Figures 3a and b]. Merely by changing the window setting (using “Adjust image window” icon on top of the window) of the FLAIR VRT images, one can mark the skin incision over the scalp as per the region of the head [Figure 3c]. Once the brain is exposed in the operation room (after raising the scalp flap, craniotomy, and dural opening), a VRT image with a lesion marked on it should be used as a guide, which resembles surface cuts of FLAIR images [Figure 4a] and exactly matches with the morphological appearance of gyri and sulci [Figures 4b and c]. One can use it for accessing the lesion over the surface or under the cortical surface.[8]
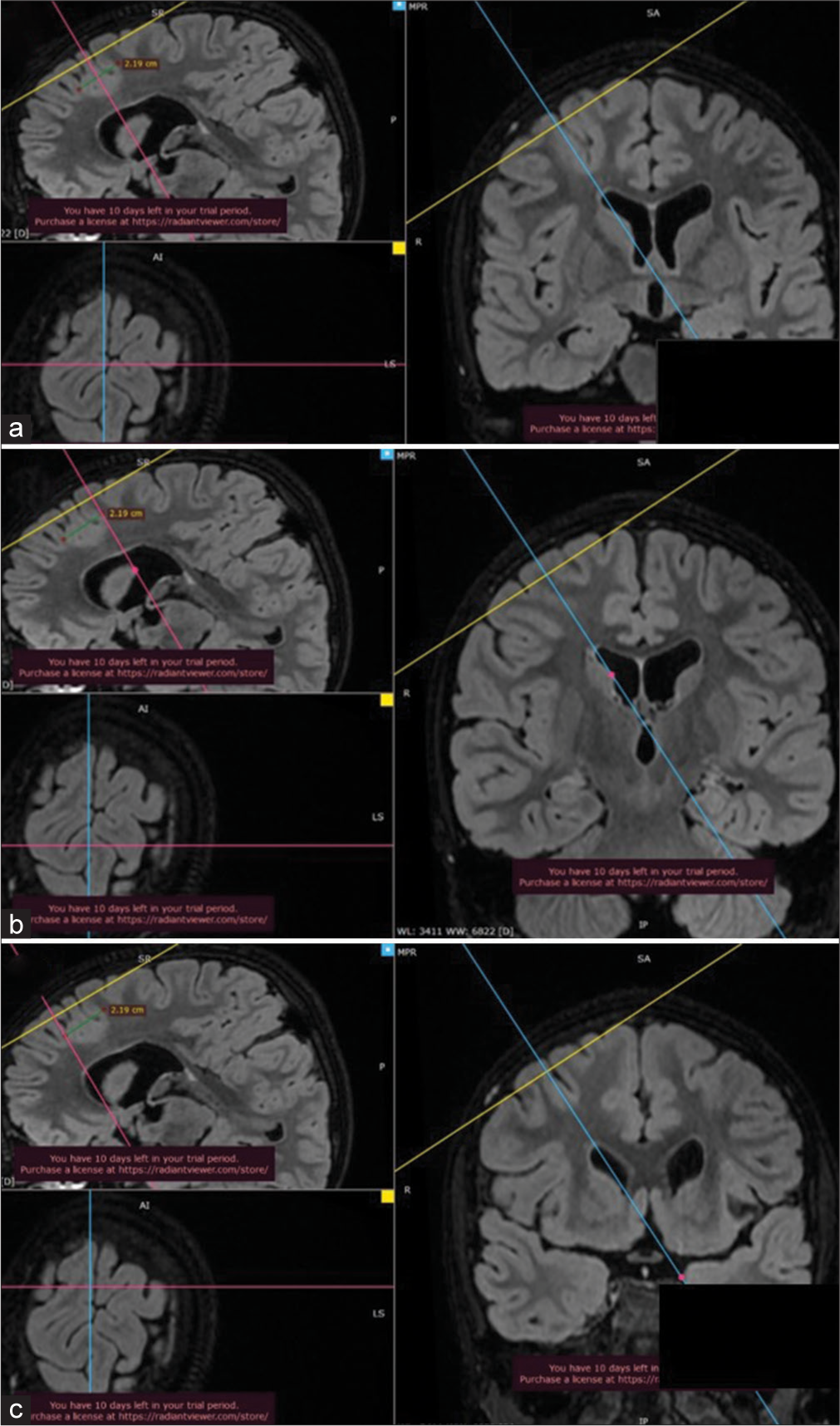
- 3D multiplanar reformation images of 3D fluid-attenuated inversion recovery magnetic resonance imaging sequences showing (a) hyper-intensity suggestive of focal cortical dysplasia in the right frontal lobe in which sagittal plane crossing through the center, (b) posterior, and (c) anterior limits of the lesion where the axial plane is showing the surface pattern of the brain with sulci and gyri.
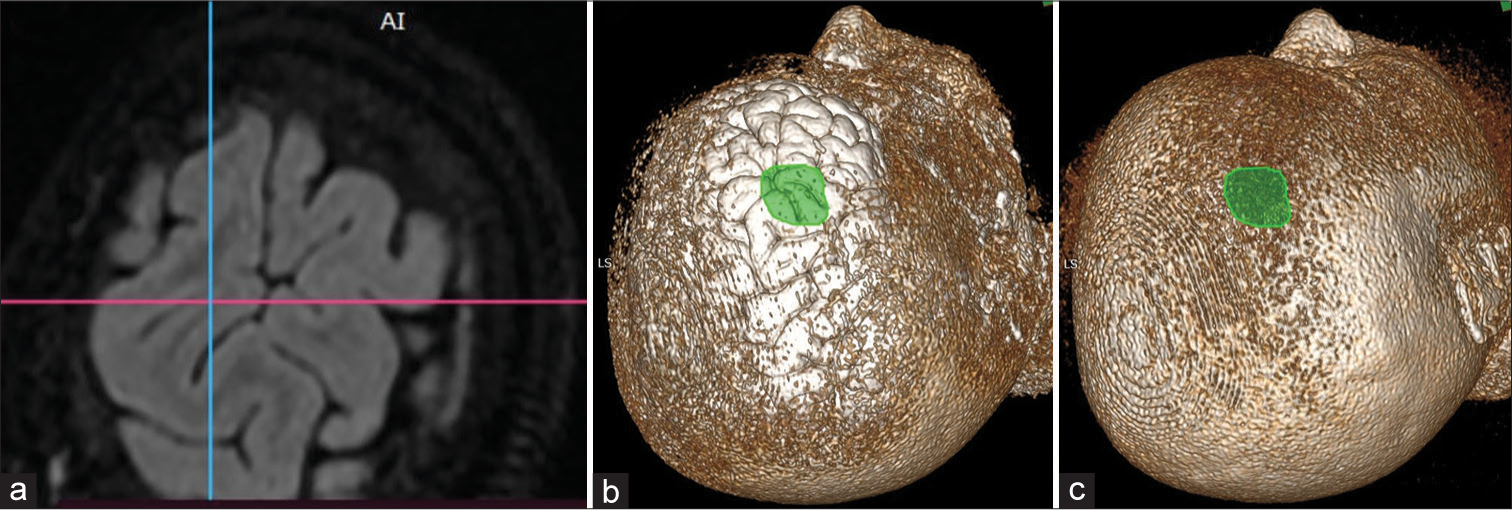
- 3D multiplanar reformation image where (a) axial plane showing a surface pattern of the brain with sulci and gyri, (b) 3D volume-rendering technique (VRT) localization of lesion (area marked in green) over similar appearing gyri and sulci of axial T2 fluid-attenuated inversion recovery, and (c) corresponding 3D VRT image of the scalp with a lesion (green area) projected on it.
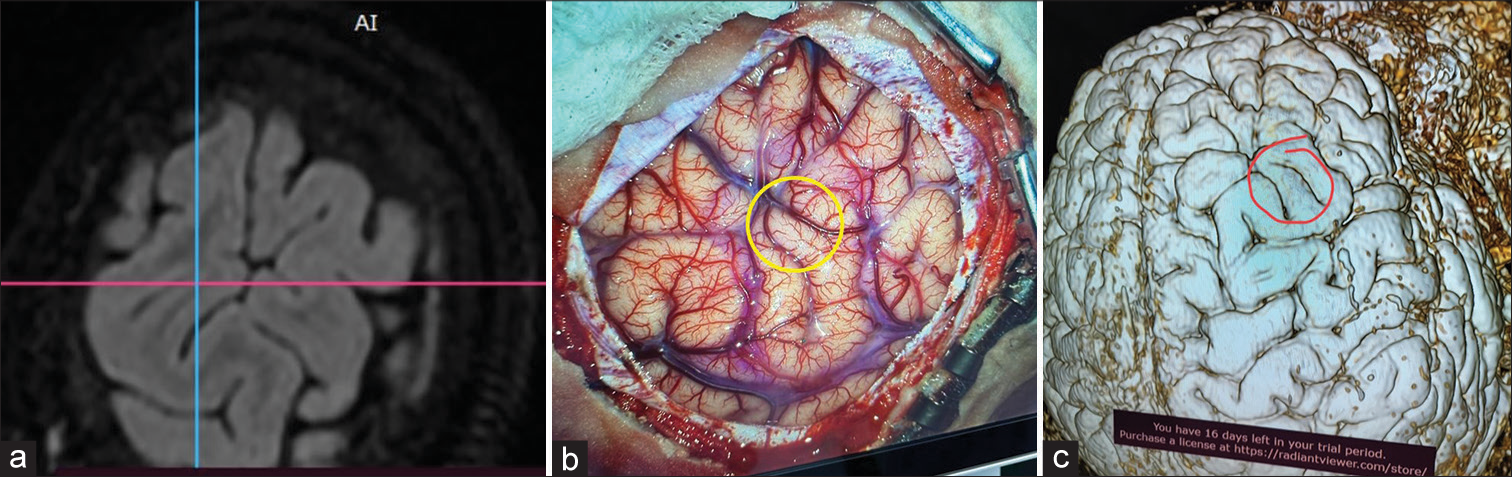
- (a) Axial image of 3D multiplanar reformation fluid-attenuated inversion recovery (FLAIR) sequence of surface, (b) exposed brain over the area with lesion marked in a yellow circle, (c) where corresponding redline lesion area is marked similar to 3D volume rendering technique FLAIR image of the brain.
The T2 3D FLAIR sequences can be used for patients above 8–10 years only to see brain gyri and sulci. Below eight years, due to density and molecular structure, brain gyri and sulci are not visible. However, 3D VRT images of CE images do show CE lesions and arteries and veins.
DISCUSSION
Clinical findings based localization is insufficient and is based on surgeons’ clinical skills. Craniometric localizations are more reliable than clinical findings alone; however, technological advancements, despite their immense applications in localization, are gradually being forgotten and are used by very few. Various types of imaging studies, the use of bony landmarks along with imaging studies, are still being used in resource-constrained settings. Neuronavigation, robotics, virtual, and augmented reality techniques, though very effective, are beyond the reach of the majority of patients of LMICs.[1-7,9,10]
A cost-effective online available software, which can be used on laptop or desktop computers, is accessible nowadays in any part of the world. High-quality MRI equipments too are nowadays available even in tier 2 cities and most of the LMICs. Using computer software is much easier for the current generation of young surgeons. The most of such softwares are quite user-friendly and need some practice for a few days only to be effectively used for surgical assistance. Authors have been using radiant DICOM software for the last many years for spine, craniovertebral junction, neurovascular, and neurooncological surgeries.[8,11,12] Readers may also refer to the report, which describes the technique for using this software.[8]
Axial, sagittal, and coronal images do help us to understand the location of the lesion, but it becomes difficult to visualize a 3D morphology of the lesion by looking at uniplanar or MPR images. Morphological appearance of the brain in the surgical field along with the lesion placed in it can be the most effective way to help a surgeon, which cannot be done even by neuronavigation or stereotactic frames. Google Lens with AR and VR techniques will definitely grow further but that too uses imaging studies, which are being used for assessing the lesion, which again is not morphological appearance.[7,9,10]
A CE MRI brain is capable of adding information before surgery about vascular anatomy over the brain surface; however, one radiant DICOM does not show both brain surface (seen in 3D FLAIR sequence) and vessels (seen in 3D CEMRI sequence) together in any sequence. One has to visualize vessels relationship on the sulci seen in the FLAIR VRT images. Once the brain surface is exposed, one can easily match the operative field appearance with the lesion marked by the VRT FLAIR images to guide the surgeon. Radiant DICOM software is for Windows only; however, a software application “HOROS” has more or less similar features and runs on Mac OS X.
Deep-seated contrast-enhancing lesions can be localized using this technique, which has been reported by the senior author.[11] This report focuses mainly on non-contrast enhancing lesions, especially over the cortical surface. The technique can be used for deeper lesions too using the 3D T2 FLAIR sequence where scalp incision, craniotomy, and entry point over the gyrus/ sulcus along with the trajectory can be decided. The VRT technique using computed tomography (CT) scan cannot be used to see the brain surface; however, its applications are useful for selected other aspects of neurosurgical planning.[8]
CONCLUSION
A cost-effective solution of software application for lesion localization in neurosurgery can be accessible to the majority of neurosurgeons of LMICs for their patients. It is an effective way to simulate a real brain-like surface with a lesion over it, which matches exactly the brain of the patients seen during surgery.
Ethical approval
Institutional Review Board approval is not required.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Craniomapper for accurate localization of lesion during craniotomy: How much benefit does it have over anatomical marking? Report of two cases. J Neurosci Rural Pract. 2014;5:202-3.
- [CrossRef] [PubMed] [Google Scholar]
- The return of the lesion for localization and therapy. Brain. 2023;146:3146-55.
- [CrossRef] [PubMed] [Google Scholar]
- Localization of intracranial lesions using superficial stereotaxic cranial lesion locator based on magnetic resonance images. Surg Innov. 2019;26:82-5.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of the reliability of brain lesion localization when using traditional and stereotactic image-guided techniques: A prospective study. J Neurosurg. 2005;103:424-7.
- [CrossRef] [PubMed] [Google Scholar]
- A novel laser-based stereotactic localization device for intracranial mass resection. Brain Hemorrhages. 2021;2:106-10.
- [CrossRef] [Google Scholar]
- Intraoperative localization of subcortical brain lesions. Acta Neurochir (Wien). 2008;150:537-42.
- [CrossRef] [PubMed] [Google Scholar]
- Use of google glass to enhance surgical education of neurosurgery residents: “Proof-of-concept” study. World Neurosurg. 2017;98:711-4.
- [CrossRef] [PubMed] [Google Scholar]
- Three-dimensional volume rendering: An underutilized tool in neurosurgery. World Neurosurg. 2019;130:485-92.
- [CrossRef] [PubMed] [Google Scholar]
- Holographic mixed-reality neuronavigation with a head-mounted device: Technical feasibility and clinical application. Neurosurg Focus. 2021;51:E22.
- [CrossRef] [PubMed] [Google Scholar]
- Intra-operative applications of augmented reality in glioma surgery: A systematic review. Front Surg. 2023;10:1245851.
- [CrossRef] [PubMed] [Google Scholar]
- Simulation of surgery for supratentorial gliomas in virtual reality using a 3D volume rendering technique: A poor man's neuronavigation. Neurosurg Focus. 2021;51:E23.
- [CrossRef] [PubMed] [Google Scholar]
- Volume rendering technique (VRT) for planning and learning cranio-vertebral junction (CVJ) surgeries: Technical note. Turk Neurosurg. 2021;31:807-12.
- [CrossRef] [PubMed] [Google Scholar]






