Translate this page into:
Prognostic Impact of the Combination of MGMT Methylation and TERT Promoter Mutation in Glioblastoma
Thara Tunthanathip Division of Neurosurgery, Department of Surgery, Faculty of Medicine, Prince of Songkla University Songkhla 90110 Thailand tsus4@hotmail.com
This article was originally published by Thieme Medical and Scientific Publishers Pvt. Ltd. and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Abstract
Background The concept of combinational analysis between the methylation of O6-methylguanine-DNA methyltransferase (MGMT) and telomerase reverse transcriptase promoter (pTERT) mutation in glioblastoma (GBM) has been reported. The main study objective was to determine the prognosis of patients with GBM based on MGMT/pTERT classification, while the secondary objective was to estimate the temozolomide effect on the survival time of GBM with MGMT/pTERT classification.
Methods A total of 50 GBM specimens were collected after tumor resection and were selected for investigating MGMT methylation and pTERT mutation. Clinical imaging and pathological characteristics were retrospectively analyzed. Patients with MGMT/pTERT classification were analyzed using survival analysis to develop the nomogram for forecasting and individual prognosis.
Results All patients underwent resection (total resection: 28%, partial resection: 64%, biopsy: 8%). Thirty-two percent of all cases received adjuvant temozolomide with radiotherapy. Sixty-four percent of the case was found methylated MGMT, and 56% of the present cohort found pTERT mutation. Following combinational analysis of biomarkers, results showed that the GBMs with methylated MGMT and wild-type pTERT had a superior prognosis compared with other subtypes. Using Cox regression analysis with multivariable analysis, the extent of resection, postoperative chemoradiotherapy, MGMT/pTERT classification were associated with a favorable prognosis. Hence, a web-based nomogram was developed for deploying individual prognostication.
Conclusions The interaction of MGMT methylation and pTERT mutation was confirmed for predicting prognosis. The results from the present study could help physicians create treatment strategies for GBM patients in real-world situations.
Keywords
telomerase reverse transcriptase
O6-methylguanine-DNA methyltransferase
primary glioblastoma
survival
prognosis
Introduction
Various biomarkers have been used as prognostic factors in malignant brain tumors, including glioblastoma (GBM). GBM is categorized into two groups by isocitrate dehydrogenase (IDH) mutation.1 In prior studies, the IDH-wild-type tumor had an inferior prognosis than the IDH-mutant tumor. In addition, IDH mutation also is one of the key biomarkers that explain the pathogenesis of gliomas.1 2
IDH-mutant GBMs have been found in just 8 to 10% of cases. Therefore, previous studies have identified various biomarkers that were prognostic factors in the majority of GBMs. For example, the mutation of the telomerase reverse transcriptase promoter (pTERT) has been reported as a prognostic factor in numerous cancers.1 3 Simon et al conducted an exploratory study in 192 patients with GBM and reported that GBM patients with pTERT mutations were found in 80.3% and significantly related to poor prognosis,4 while Kim et al found pTERT mutation in 59.2% (25/42) of all GBM cases.5
In addition, methylation of O6-methylguanine-DNA methyltransferase (MGMT) has been studied. The epigenetic methylation predicted the responsiveness of alkylating agents. Esteller et al found that glioma with methylation of MGMT interfered with MGMT protein-encoding. Therefore, chemotherapy significantly affected the tumor cells. Gliomas with methylated MGMT (mMGMT) are called chemosensitivity tumors, while chemo-resistant tumors are gliomas with unmethylated MGMT (uMGMT), which create O6-methylguanine-DNA methyltransferase to change the structure of the alkylating agents and resist the chemotherapy.6 Hegi et al studied the association between mMGMT and prognosis in patients with GBM. They advised that mMGMT was a significantly favorable predictor. Moreover, patients with mMGMT who received temozolomide (TMZ) with radiotherapy (RT) had a significantly longer median survival time than patients who received RT alone.7 Recently, the National Comprehensive Cancer Network has proposed the clinical practice guideline for central nervous system tumor (CNS) treatment. Methylation of MGMT was considered for selecting the postoperative management in GBM.8
The concept of a combined analysis of biomarkers for predicting prognosis in GBM patients has been proposed in literature reviews. Arita et al studied 453 GBM cases and found the interaction between the methylation of MGMT and pTERT mutation related to prognosis. In detail, GBMs were categorized into four subgroups as follows: methylated MGMT with wild-type TERT (mMGMT-wTERT), methylated MGMT with mutant TERT (mMGMT-wTERT), uMGMT with mutant TERT (uMGMT-wTERT), uMGMT with wild-type TERT (uMGMT-mTERT), called MGMT/pTERT classification. Hence, uMGMT-mTERT GBMs had the poorest prognosis, while mMGMT-wTERT and mMGMT-mTERT GBMs had the most favorable prognosis. Additionally, uMGMT-wTERT GBMs had an average prognosis.9 Therefore, several prior studies found similar interactions between MGMT promoter methylation and TERT mutation.10 11 12 However, a lack of evidence was mentioned concerning the effect of TMZ on survival in MGMT/pTERT classification. The present study aimed to estimate the prognosis of patients with GBM based on MGMT/pTERT classification. Also, the secondary objective was to estimate the TMZ effect on the survival time of GBM with MGMT/pTERT classification.
Methods
Study Design and Population
The Ethics Committee and Institutional Review Board of the Faculty of Medicine, Prince of Songkla University, reviewed and approved the present study (REC 61–065–10–1). All cases were diagnosed as GBMs between 2003 and 2018, and histological slides were reviewed to confirm the diagnosis. Electronic medical records were reviewed for clinical data collection. Moreover, preoperative and postoperative imaging of GBM patients was evaluated by a neurosurgeon.
DNA Extraction and Molecular Analysis
DNA was extracted according to the manufacturer's instructions. Mutation of IDH1 and methylation of MGMT were investigated, as formerly described.13 In detail, the methylated MGMT was defined as 30% or more methylation.14 pTERT mutations were identified by droplet digital polymerase chain reaction with C228T_113 Assay (Bio-Rad; Assay ID; dHsaEXD72405942) based on the study of Corless et al.15
One-hundred and seventy-three patients were treated between January 2003 and December 2018. Therefore, the GBM patients who did not have tumor specimens for the molecular study were excluded. Finally, the remaining 50 patients were completely analyzed for IDH1 mutation, MGMT methylation, and pTERT mutation.
Statistical Analysis
Descriptive statistics were performed to explain the demographic data of the present cohort. The categorical variables were reported as percentages, and mean ± standard deviation (SD) was used for the continuous variables with normal distribution. Moreover, continuous variables without normal distribution were described by the median and interquartile range (IQR).
Survival analysis was performed for estimating prognosis. The overall survival (OS) and survival probability in each time-point were analyzed. The Kaplan–Meier survival curves were visually analyzed for survival time of each variable. The Cox hazard regression was used for identifying the prognostic factors in univariate analysis and multivariable analysis. In detail, the backward stepwise method was performed to select the final predictive model. A p < 0.05 was accepted as being statistically significant. Hence, the final model following multivariable analysis was used for nomogram development, as previously described,16 17 and the nomogram was deployed as a web-based application for general practice in the future.
After nomogram development, the nomogram scoring system was used to estimate the predictive performance, including discrimination, calibration, and internal validation.18 In detail, Harrell's concordance index (C-index) was estimated for discrimination by Cox hazard regression, and calibration was visually evaluated by closing a 45-degree line. Additionally, internal validation was conducted by the bootstrap validation with 200-time resampling, and the result of the validation was reported as the area under the ROC curves (AUC).19
The statistical analysis was performed using the R program version 3.4.0 software (R Foundation, Vienna, Austria). Moreover, a web-based nomogram was developed through https://www.shinyapps.io/.
Results
Clinical Characteristics
Fifty patients with GBM were described with baseline clinical characteristics (Table 1). Of them, 58% were male with a median age of 54 years old (IQR: 23). The common signs and symptoms of patients were progressive headache, hemiparesis, and seizure. In addition, the majority of GBM were single and involved the frontal lobe. In addition, more than half of the tumors infiltrated the eloquent area defined according to Lacroix et al.20
|
Factors |
n (%) |
|---|---|
|
Age, years |
|
|
< 60 |
30 (60.0) |
|
≥ 60 |
20 (40.0) |
|
Median age-year (IQR) |
54 (23) |
|
Gender |
|
|
Male |
29 (58.0) |
|
Female |
21 (42.0) |
|
Signs and symptoms |
|
|
Progressive headache |
26 (52.0) |
|
Motor weakness |
20 (40.0) |
|
Seizure |
14 (28.0) |
|
Alteration of consciousness |
9 (18.0) |
|
Behavior changes |
6 (12.0) |
|
Aphasia |
3 (6.0) |
|
Ataxia |
1 (2.0) |
|
Preoperative Karnofsky performance status |
|
|
< 80 |
22 (44.0) |
|
≥80 |
28 (56.0) |
|
Number of tumor |
|
|
Single |
48 (96.0) |
|
Multiple |
2 (4.0) |
|
Tumor location |
|
|
Eloquent areaa |
27 (54.0) |
|
Frontal lobe |
19 (38.0) |
|
Temporal lobe |
12 (24.0) |
|
Corpus callosum |
8 (16.0) |
|
Parietal lobe |
|
|
Occipital lobe |
4 (8.0) |
|
Basal ganglion |
3 (6.0) |
|
Suprasellar area |
|
|
Leptomeningeal dissemination |
|
|
No |
43 (86.0) |
|
Brain |
5 (10.0) |
|
Brain with spinal cord |
2 (4.0) |
|
Mean diameter of tumor-cm (SD) |
5.20 (1.72) |
|
Extent of resection |
|
|
Neuronavigator-guided biopsy |
4 (8.0) |
|
Partial resection |
32 (64.0) |
|
Total resection |
14 (28.0) |
|
Postoperative KPS |
|
|
< 80 |
31 (62.0) |
|
≥80 |
19 (38.0) |
|
Adjuvant treatment |
|
|
No adjuvant treatment |
17 (34.0) |
|
Radiotherapy alone |
17 (34.0) |
|
Temozolomide with radiotherapy |
16 (32.0) |
|
IDH1 mutation |
|
|
Wild-type IDH1 |
46 (92.0) |
|
Mutant IDH1 |
4 (8.0) |
|
MGMT methylation |
|
|
Unmethylated MGMT |
18 (36.0) |
|
Methylated MGMT |
32 (64.0) |
|
TERT promoter mutation |
|
|
Wild-type TERT |
22 (44.0) |
|
Mutant TERT |
28 (56.0) |
|
Combined biomarkers |
|
|
Methylated MGMT with wild-type TERT |
18 (36.0) |
|
Methylated MGMT with mutant TERT |
14 (28.0) |
|
Unmethylated MGMT with wild-type TERT |
14 (28.0) |
|
Unmethylated MGMT with mutant TERT |
4 (8.0) |
Abbreviations: GBM, glioblastoma; IDH1, isocitrate dehydrogenase1; IQR, interquartile range; KPS, Karnofsky Performance Status; MGMT, O6-methylguanine-DNA methyltransferase; RT, radiotherapy; SD, standard deviation; TERT, telomerase reverse transcriptase; TMZ, temozolomide.
All patients underwent surgical management. Total tumor resection was performed in 28% of the present cohort. One-third of the cohort refused any further treatment following surgery. TMZ is used postoperatively in 32% of cases in real-world situations, primarily because patients cannot afford the cost of TMZ.
In biomarker profiles, the status of IDH1 was almost all IDH-wild-type GBMs, whereas mMGMT was found in 64% of cases. In addition, pTERT methylation was frequently found in 56% of cases. When GBM was categorized based on the status of MGMT methylation and pTERT mutation, tumors were divided into four subgroups as follows: mMGMT-wTERT, mMGMT-mTERT, uMGMT-wTERT, and uMGMT-mTERT GBMs. The most common subgroup was mMGMT-wTERT GBM, found in 36.0% of cases.
Survival Analysis
The mean follow-up time was 13.17(SD: 9.6) months, and the median OS of the present study was 11 months (95% confidence interval [CI]: 9.15–12.84), as shown in Fig. 1A. Additionally, 1, 2, and 3-year survival probabilities were 30.8, 12.6, and 0.42%, respectively. When the Kaplan–Meier survival analysis was performed on each variable, the total tumor resection, adjuvant therapy with TMZ and RT, mMGMT-wTERT tumor significantly prolonged survival time, as shown in Fig. 1B–F.
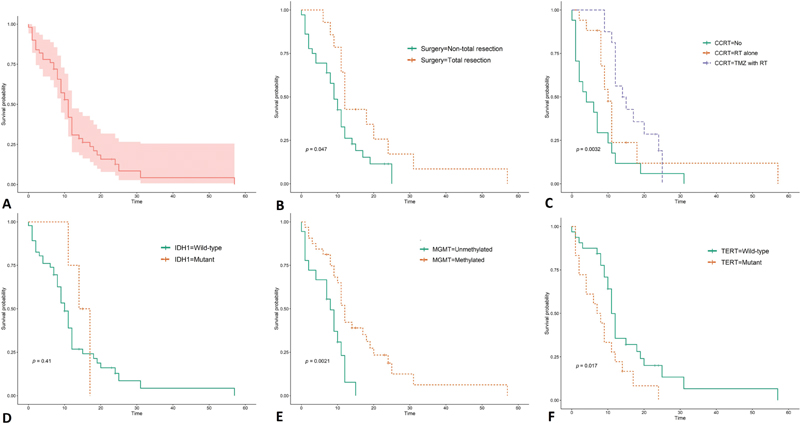
-
Fig. 1 Survival curves of patients with glioblastoma. (A) Overall, (B) extent of resection, (C) concurrent chemoradiotherapy (CCRT), (D) isocitrate dehydrogenase (IDH1) mutation, (E) O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation, and (F) telomerase reverse transcriptase (TERT) promoter mutation. TMZ, temozolomide.
Fig. 1 Survival curves of patients with glioblastoma. (A) Overall, (B) extent of resection, (C) concurrent chemoradiotherapy (CCRT), (D) isocitrate dehydrogenase (IDH1) mutation, (E) O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation, and (F) telomerase reverse transcriptase (TERT) promoter mutation. TMZ, temozolomide.
According to the combined MGMT/TERT classification, mMGMT-wTERT GBMs had a favorable prognosis with a median OS of 18 months (95% CI: 10.3–25.6), while uMGMT-mTERT GBMs had the shortest survival time of 1 month, as shown in Fig. 2. Moreover, the median OS of mMGMT-mTERT and uMGMT-mTERT were 9 (95% CI: 7.1–10.8) and 9 months (95% CI: 6.7–11.2), respectively.
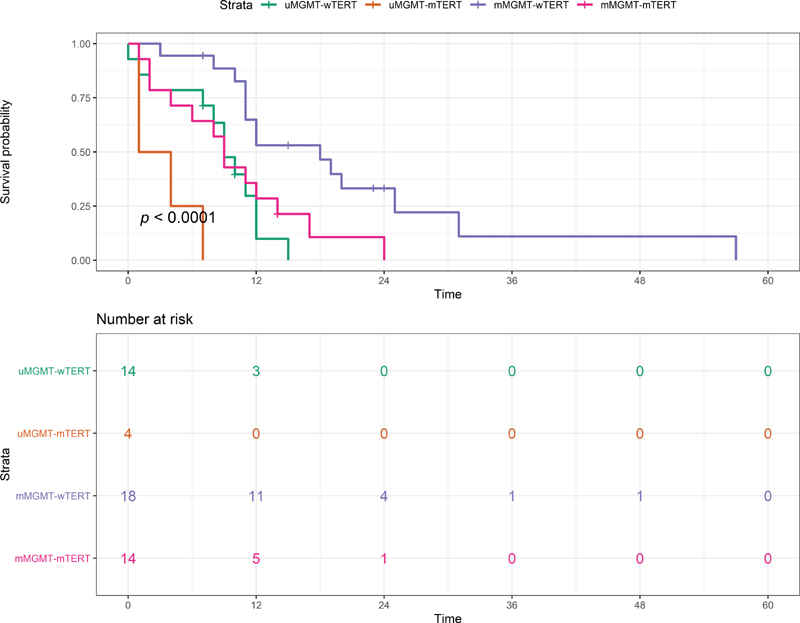
-
Fig. 2 Survival curves of the O6-methylguanine-DNA methyltransferase/promotor telomerase reverse transcriptase (MGMT/pTERT) classification. mMGMT, methylated O6-methylguanine-DNA methyltransferase; mTERT, methylated telomerase reverse transcriptase; uMGMT, unmethylated O6-methylguanine-DNA methyltransferase; wTERT, wild-type telomerase reverse transcriptase.
Fig. 2 Survival curves of the O6-methylguanine-DNA methyltransferase/promotor telomerase reverse transcriptase (MGMT/pTERT) classification. mMGMT, methylated O6-methylguanine-DNA methyltransferase; mTERT, methylated telomerase reverse transcriptase; uMGMT, unmethylated O6-methylguanine-DNA methyltransferase; wTERT, wild-type telomerase reverse transcriptase.
We also analyzed the survival impact of TMZ with RT based on the combined MGMT/TERT classification, as shown in Table 2. TMZ with RT significantly benefited mMGMT-wTERT GBMs with a median OS of 25 months, whereas other subgroups had a median OS of 12 to 14 months (log-rank test, p = 0.03). For subgroup analysis of patients who received TMZ with RT, patients with mMGMT seemed to have survival benefit from concurrent chemoradiotherapy.
|
The combined MGMT/TERT classification |
Median survival time (95% CI)a |
Hazard ratio (95% CI) |
|---|---|---|
|
Methylated MGMT with wild-type TERT (n = 5) |
25 (10.3–25.6) |
Ref |
|
Methylated MGMT with mutant TERT (n = 7) |
14 (8.8–19.1) |
4.5 (0.84–23.5) |
|
Unmethylated MGMT with wild-type TERT (n = 0) |
12 (9.4–14.5) |
8.5 (1.2–56.8) |
|
Unmethylated MGMT with mutant TERT (n = 4) |
– |
– |
Abbreviations: CI, confidence interval; GBM, glioblastoma; MGMT, O6-methylguanine-DNA methyltransferase; TERT, telomerase reverse transcriptase.
For prognostication, Cox hazard regression was performed. The extent of resection, TMZ with RT, and MGMT/TERT classification were the potential prognostic models for both univariate and multivariable analysis, as shown in Table 3.
|
Univariate analysis |
Multivariable analysis |
|||
|---|---|---|---|---|
|
Factors |
Hazard ratio (95% CI) |
p-Value |
Hazard ratio (95% CI) |
p-Value |
|
Age, years |
||||
|
< 60 |
Ref |
|||
|
≥60 |
1.59 (0.86–2.94) |
0.13 |
||
|
Gender |
||||
|
Male |
Ref |
|||
|
Female |
||||
|
Preoperative Karnofsky performance status |
||||
|
< 80 |
Ref |
|||
|
> 80 |
0.70 (0.38–1.29) |
0.25 |
||
|
Tumor location |
||||
|
1.44 (0.78–2.67) |
0.23 |
|||
|
Temporal lobea |
1.10 (0.53–2.27) |
0.78 |
||
|
Corpus callosuma |
1.17 (0.51–2.65) |
0.70 |
||
|
Frontal lobea |
0.59 (0.31–1.14) |
0.11 |
||
|
Parietal lobea |
1.58 (0.69–3.62) |
0.27 |
||
|
Occipital lobea |
0.59 (0.17–1.99) |
0.39 |
||
|
Diameter of maximum tumor, cm |
||||
|
< 5 |
Ref |
|||
|
> 5 |
0.90 (0.48–1.66) |
0.90 |
||
|
Extent of resection |
||||
|
Nontotal resection |
Ref |
Ref |
||
|
Total resection |
0.51 (0.25–0.99) |
0.05 |
0.44 (0.20–0.98) |
0.05 |
|
Postoperative Karnofsky performance status |
||||
|
< 80 |
Ref |
|||
|
> 80 |
0.54 (0.28–1.03) |
0.06 |
||
|
Adjuvant treatment |
||||
|
No adjuvant treatment |
Ref |
Ref |
||
|
Radiotherapy alone |
0.50 (0.23–1.05) |
0.06 |
0.57 (0.24–1.30) |
0.18 |
|
Temozolomide with radiotherapy |
0.31 (0.14–0.65) |
0.002 |
0.12 (0.04–0.35) |
<0.001 |
|
IDH1 mutation |
||||
|
Wild-type IDH1 |
Ref |
|||
|
Mutant IDH1 |
0.62 (0.19–2.00) |
0.44 |
||
|
MGMT promoter methylation |
||||
|
Methylated MGMT |
Ref |
|||
|
Unmethylated MGMT |
0.37 (0.18–0.73) |
0.004 |
||
|
TERT promoter mutation |
||||
|
Wild-type TERT |
Ref |
|||
|
Mutant TERT |
2.22 (1.14–4.35) |
0.01 |
||
|
Combined biomarkers |
||||
|
Methylated MGMT with wild-type TERT |
Ref |
Ref |
||
|
Methylated MGMT with mutant TERT |
2.91 (1.27–6.65) |
0.01 |
9.52 (3.29–27.53) |
<0.001 |
|
Unmethylated MGMT with mutant TERT |
3.74 (1.54–9.08) |
0.004 |
6.99 (2.55–19.15) |
<0.001 |
|
Unmethylated MGMT with wild-type TERT |
16.99 (4.47–64.60) |
<0.001 |
14.38 (3.43–60.27) |
<0.001 |
Abbreviations: CI, confidence interval; IDH1, isocitrate dehydrogenase1; MGMT, O6-methylguanine-DNA methyltransferase; TERT, telomerase reverse transcriptase.
Patients who underwent an operation with total tumor removal had a significantly better prognosis than nontotal tumor removal (hazard ratio [HR]: 0.44, 95% CI: 0.20–0.98). For adjuvant therapy, TMZ treatment had the benefit of prolonging survival time (HR: 0.12, 95% CI: 0.04–0.35). Against a reference of mMGMT-wTERT GBMs, other subgroups had a significantly inferior prognosis than the reference. For implication in general practice, we developed the nomogram, a two-dimensional graphic scoring system for individual 1, 2, and 3-year prognostication, as shown in Fig. 3. Moreover, the web-based nomogram was deployed for ease to predict an individual in general practice via https://thara.shinyapps.io/Nomogram_GBM_PSU/; the code for the prediction tools was linked through https://github.com/Thara-PSU/Nomogram_GBM_PSU.
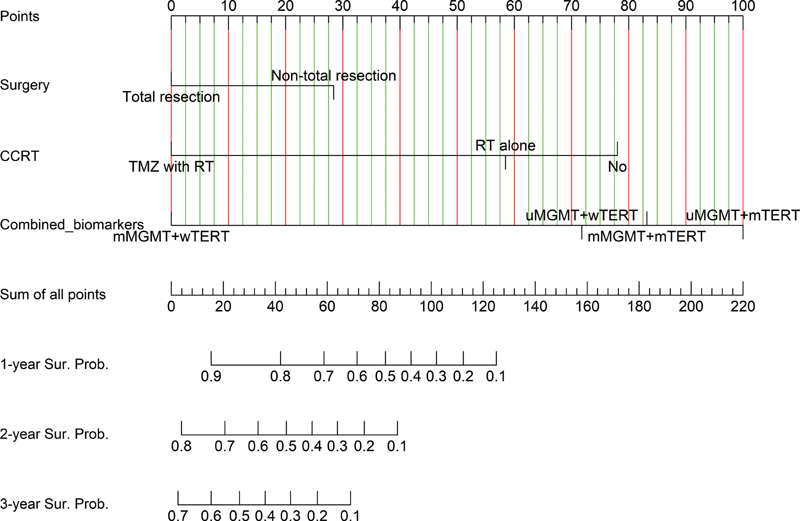
-
Fig. 3 Nomogram for predicting the individual survival of glioblastoma patient. CCRT, concurrent chemoradiotherapy; mMGMT, methylated O6-methylguanine-DNA methyltransferase; mTERT, methylated telomerase reverse transcriptase; TMZ, temozolomide; uMGMT, unmethylated O6-methylguanine-DNA methyltransferase; wTERT, wild-type telomerase reverse transcriptase.
Fig. 3 Nomogram for predicting the individual survival of glioblastoma patient. CCRT, concurrent chemoradiotherapy; mMGMT, methylated O6-methylguanine-DNA methyltransferase; mTERT, methylated telomerase reverse transcriptase; TMZ, temozolomide; uMGMT, unmethylated O6-methylguanine-DNA methyltransferase; wTERT, wild-type telomerase reverse transcriptase.
For evaluation of the predictive nomogram, Harrell's C-index was 0.791 for the model discrimination and the calibration plot was constructed, as shown in Fig. 4. Moreover, bootstrap validation was performed for the internal validation, wherein the AUC was 0.79.
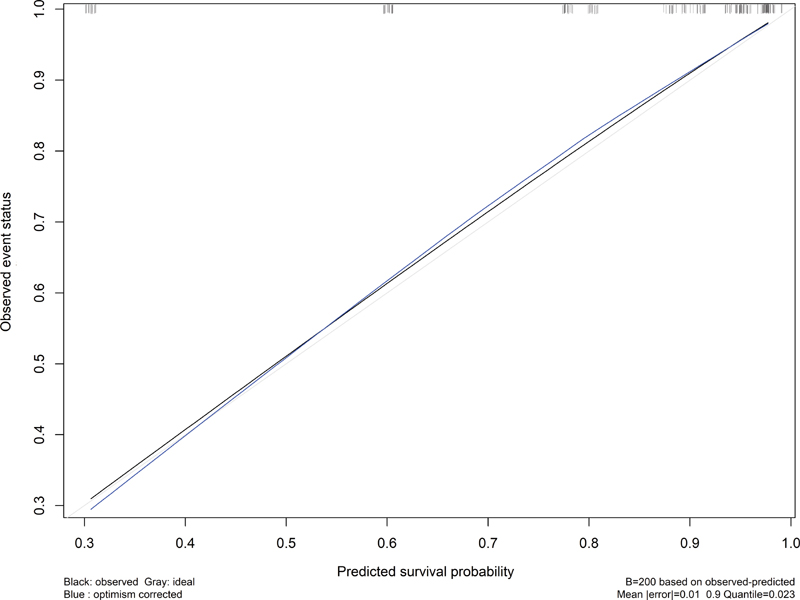
-
Fig. 4 Calibration plot of the nomogram closing the 45-degree line.
Fig. 4 Calibration plot of the nomogram closing the 45-degree line.
Discussion
In the present study, we found the key biomarkers that significantly predicted prognosis, while the interaction of MGMT methylation status and pTERT mutation was also detected. The unmethylation of MGMT promoter and mutation of TERT influenced the shortened survival time in the present study. Therefore, patients with mMGMT-wTERT GBM had the longest survival time, while the poorest prognosis was for patients with uMGMT-mTERT GBM. These findings were in accordance with prior studies. Purkait et al stratified the subgroups of GBMs based on both biomarkers and reported that patients with uMGMT-mTERT GBM had the shortest survival time (HR: 11.12, 95% CI: 1.99–61.99).10 Similarly, Sasaki et al studied these biomarkers for predicting prognosis in elderly GBM patients and found that the poorest prognosis was the uMGMT-mTER subtype.12
According to the CNS tumor classification, GBMs are categorized by IDH mutation.1 The wild-type IDH1 GBMs were commonly found in the primary GBM and associated with poor prognosis.1 2 3 However, IDH1 mutation was not an independent prognostic factor in the present study. One prior meta-analysis study reported no association of IDH1 mutation in eastern studies when subgroup analysis was performed based on country region. The effect of IDH1 mutation on prognosis is likely related to the topographic connection.21 This biomarker should be explored for its association with prognostication in future studies.
We observed the three key prognostic factors through a multivariable Cox regression analysis: the extent of resection, CCRT, and MGMT/pTERT classification. Currently, TMZ with RT is the standard treatment following tumor resection.22 23 However, the cost of TMZ is high and becomes an economic burden in a limited-resource setting. Therefore, selecting GBM patients by predicted favorable prognosis has been proposed for the likelihood of TMZ cost-effectiveness in real-world situations.24
Nomogram is one of the clinical prediction tools (CPT) that have been used for various diseases such as cancer,25 trauma,26 27 or degenerative disease.28 Therefore, we proposed a web-based nomogram for prognostication that will be applied for selecting patients with a favorable prognosis when low- or middle-income settings need to allocate resources.
According to the concept of translational medicine, T0 research defines the phase of basic science research studied to explore the novel biomarkers or candidate genes through -omic technologies. Further, T1 research defines research translated to humans that involves the tools for screening or diagnostic testing.29 30 Sam et al studied genetic polymorphism and the risk factors for screening gastrointestinal tract carcinoma in Indian people.31 Hence, T2 research translates information to patients as an evidence-based guideline that commonly studies cancer staging, prognosis, and treatment response prediction. In detail, the development of CPT corresponds with development, calibration, discrimination, and internal validation in the derivation of CPT.32 The T3 research translates to general practice via dissemination studies. Therefore, this is in accordance with the external validation phase in the CPT development, while the impact study of the tools achieved T4 research impacts to the society and community as well as changing the health policy or economics.33
The present study developed and proposed a nomogram for predicting prognosis by integrating clinical variables and biomarkers. Therefore, the prediction performance of the nomogram was estimated in steps as the T2 research of translational medicine. Harrell's C-index and AUC of bootstrap validation in the present study were acceptable.34 35 Therefore, the line of the calibration plot in the present study was close the 45-degree line that accepted the performance. The nomogram developed from the GBM patients will be evidence to conduct external validation or dissemination study to apply in health practice in the future.
Nevertheless, certain limitations should also be recognized. First, we considered a small sample of GBM patients. Multicenter research or systematic review and meta-analysis will potentially confirm this association by cumulative study populations. Second, the investigation of pTERT mutation was performed only at positions -124 (C228T); pTERT mutation at another position may have been missed. However, we used droplet digital polymerase chain reaction assays for detecting the mutations, a technique that has the potential to detect pTERT mutations15 36 specifically. For future research, a multicenter study should be conducted to increase the sample size as well as to confirm the association of MGMT/pTERT classification and prognosis in GBMs. Moreover, comparison between nomograms and other prediction tools such as clinical prediction rules37 or machine learning algorithms17 should be performed to evaluate their predictive performance.
Conclusion
The interaction of MGMT methylation and pTERT mutation was confirmed for predicting prognosis. The results from the present study could help physicians create treatment strategies for GBM patients in real-world situations.
Acknowledgment
The authors would like to offer their special thanks to Associate Professor Paramee Thongsuksai and Dr. Pasuree Sangsupawanich for their advice about manuscript preparation. Furthermore, they also thank Mrs. Wanwisa Maneechay and Ms. Kanda Tongmit for administration about direct sequencing and immunohistochemistry investigation, respectively.
Conflict of Interest
None declared.
Funding The study was supported by the Faculty of Medicine, Prince of Songkla University (Kho Hong, Thailand; grant no. 61-065-10-1).
References
- The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803-820.
- [Google Scholar]
- An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807-1812. (5897):
- [Google Scholar]
- WHO Classification of Tumors of the Central Nervous System. (4th edition). Lyon: IARC Press; 2016.
- [Google Scholar]
- TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro-oncol. 2015;17(1):45-52.
- [Google Scholar]
- Clinical implications of TERT promoter mutation on IDH mutation and MGMT promoter methylation in diffuse gliomas. Pathol Res Pract. 2018;214(6):881-888.
- [Google Scholar]
- Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343(19):1350-1354.
- [Google Scholar]
- MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997-1003.
- [Google Scholar]
- Central nervous system cancers, version 3.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2020;18(11):1537-1570.
- [Google Scholar]
- A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79.
- [Google Scholar]
- Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro-oncol. 2017;19(3):394-404.
- [Google Scholar]
- Prognostic stratification of GBMs using combinatorial assessment of IDH1 mutation, MGMT promoter methylation, and TERT mutation status: experience from a tertiary care center in India. Transl Oncol. 2016;9(4):371-376.
- [Google Scholar]
- Characteristics and outcomes of elderly patients with diffuse gliomas: a multi-institutional cohort study by Kansai molecular diagnosis network for CNS tumors. J Neurooncol. 2018;140(2):329-339.
- [Google Scholar]
- The clinical characteristics and prognostic factors of multiple lesions in glioblastomas. Clin Neurol Neurosurg. 2020;195:105891.
- [Google Scholar]
- Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity in glioblastoma. J Neurooncol. 2016;128(2):333-339.
- [Google Scholar]
- Development of novel mutation-specific droplet digital PCR assays detecting TERT promoter mutations in tumor and plasma samples. J Mol Diagn. 2019;21(2):274-285.
- [Google Scholar]
- Development and validation of a nomogram for predicting the mortality after penetrating traumatic brain injury. Bull Emerg Trauma. 2019;7(4):347-354.
- [Google Scholar]
- Machine learning applications for the prediction of surgical site infection in neurological operations. Neurosurg Focus. 2019;47(2):E7.
- [Google Scholar]
- Estimation of C-index for Cox proportional hazards model with censored biomarker covariate subject to limits of detection. J Biopharm Stat. 2015;25(3):459-473.
- [Google Scholar]
- Clinical nomogram predicting intracranial injury in pediatric traumatic brain injury. J Pediatr Neurosci. 2020;15(4):409-415.
- [Google Scholar]
- A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190-198.
- [Google Scholar]
- No association between isocitrate dehydrogenase 1 mutation and increased survival of glioblastoma: a meta-analysis. PNR. 2020;11(1):1-8.
- [Google Scholar]
- Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987-996.
- [Google Scholar]
- Temozolomide for patients with wild-type isocitrate dehydrogenase (IDH) 1 glioblastoma using propensity score matching. Clin Neurol Neurosurg. 2020;191:105712.
- [Google Scholar]
- Subgroup economic analysis for glioblastoma in a health resource-limited setting. PLoS One. 2012;7(4):e34588.
- [Google Scholar]
- Prognostic factors and nomogram predicting survival in diffuse astrocytoma. J Neurosci Rural Pract. 2020;11(1):135-143.
- [Google Scholar]
- Factors associated with in-hospital mortality in severe burn patients in Songklanagarind hospital: a retrospective study. J Health Sci Med Res. 2021;x:401-410.
- [Google Scholar]
- Necessity of in-hospital neurological observation for mild traumatic brain injury patients with negative computed tomography brain scans. J Health Sci Med Res. 2020;38:267-274.
- [Google Scholar]
- A predictive model and nomogram for predicting return to work at 3 months after cervical spine surgery: an analysis from the quality outcomes database. Neurosurg Focus. 2018;45(5):E9.
- [Google Scholar]
- Translational research in cancer genetics: the road less traveled. Public Health Genomics. 2011;14(1):1-8.
- [Google Scholar]
- What are the T0 to T4 Research Classifications? [internet] . [cited 2021 May 5]. Accessed June 23, 2021 at:
- [Publisher] [Google Scholar]
- ABCB1 genetic polymorphism and risk of upper aerodigestive tract cancers among smokers, tobacco chewers and alcoholics in an Indian population. Pharmacogenet Genomics. 2007;17(10):861-866.
- [Google Scholar]
- Prognosis Research Strategy (PROGRESS) 3: prognostic model research. PLoS Med. 2013;10(2):e1001381.
- [Google Scholar]
- Framework for the impact analysis and implementation of Clinical Prediction Rules (CPRs) BMC Med Inform Decis Mak. 2011;11:62.
- [Google Scholar]
- How to develop, validate, and compare clinical prediction models involving radiological parameters: study design and statistical methods. Korean J Radiol. 2016;17(3):339-350.
- [Google Scholar]
- On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011;30(10):1105-1117.
- [Google Scholar]
- Development of sensitive droplet digital PCR assays for detecting urinary TERT promoter mutations as non-invasive biomarkers for detection of urothelial cancer. Cancers (Basel). 2020;12(12):3541.
- [Google Scholar]
- Clinical prediction rules: what are they and what do they tell us? Aust J Physiother. 2006;52(3):157-163.
- [Google Scholar]






