Translate this page into:
Neurotrophin-3 gene polymorphism in schizophrenia and its relation with diseases severity and cognitive dysfunction
*Corresponding author: Hanumanthappa Nandeesha, Department of Biochemistry, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India. nandijipmer@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Keshri N, Nandeesha H, Rajappa M, Menon V. Neurotrophin-3 gene polymorphism in schizophrenia and its relation with diseases severity and cognitive dysfunction. J Neurosci Rural Pract 2023;14:501-8.
Abstract
Objectives:
Synaptic plasticity markers are known to alter in schizophrenia. The objective of the study was to investigate the genotype and allele frequency of neurotrophin-3 (NT-3) gene polymorphism (rs6489630, rs6332, and rs11063714) and plasma NT-3 levels in schizophrenia and their relation with cognitive status.
Materials and Methods:
The study was conducted on 216 Schizophrenia patients and 216 controls. Single-nucleotide polymorphism (SNP) of NT-3 and its plasma levels were assessed in both groups. Cognitive status was evaluated using Addenbrooke Cognitive examination-III scores.
Results:
The rs6489630 polymorphism was found to be significantly associated with the severity of schizophrenia (P = 0.004). The CT genotype (P = 0.02, OR = 1.631 [1.10–2.43]) and minor allele T (P = 0.004, OR = 1.58 [1.16–2.16]) of rs6489630 conferred an increased susceptibility to develop schizophrenia. The rs6332 variant was found to affect cognitive status significantly in schizophrenia (P = 0.040), and memory dysfunction was seen in individuals with AG (P < 0.01) and AA variant (P = 0.03) of rs6332.
Conclusion:
We conclude that SNPs of NT-3 enhance the risk of schizophrenia and are related to cognitive dysfunction.
Keywords
Schizophrenia
Synaptic plasticity
Neurotrophin-3
Cognition
INTRODUCTION
Schizophrenia is a neurodevelopmental disorder affecting 20 million people worldwide and is associated with an increased risk of morbidity and mortality.[1] Cognitive impairment is one of the complications of schizophrenia, and it was attributed by earlier investigators to alteration in synaptic plasticity.[2] Cognitive rehabilitation treatment was reported to improve functional and occupational outcomes in patients with schizophrenia.[3] Previous studies have demonstrated alteration in neuroplasticity markers and its relation with cognitive dysfunction in schizophrenia.[4,5]
Neurotrophin-3 (NT-3) is a growth factor involved in the regulation of neural cell growth, differentiation, and survival.[6] NT-3 modulates the architecture of neurites leading to changes in synaptic function which is responsible for cognition, learning, behavior, and memory.[7] NT-3 is known to play a role in developing several neuropsychiatric conditions such as bipolar disorder, schizophrenia, and depression.[4,6,8] NT-3 is considered a potential candidate gene in schizophrenia and reduced mRNA levels of NT-3 have been reported by earlier studies.[9] The NT-3 gene is positioned on chromosome 12p13. Single-nucleotide polymorphisms (SNPs) of the NT-3 gene (rs6489630, rs6332, and rs11063714) were known to have functional consequences and were found to increase the risk of adolescent idiopathic scoliosis and Alzheimer’s disease.[10,11] Hattori et al have identified SNPs in the NT-3 promoter region and revealed that the genotype frequencies of the G/-3004/A polymorphism were significantly different between schizophrenics and controls.[12] However, the data related to the association of the NT-3 polymorphism with cognitive function and disease severity in schizophrenia is lacking. The primary objective of the study was to analyze the genotype and allele frequencies of NT-3 polymorphism rs6489630, rs6332, and rs11063714) between schizophrenia and controls. The secondary objective was to assess the relation of NT-3 polymorphism with cognitive scores and positive and negative symptom scores (PANSS) in schizophrenia.
MATERIALS AND METHODS
Study participants
The present study was a case–control genetic study conducted in Biochemistry and Psychiatry departments, JIPMER, Puducherry, India. Institute ethics committee approval was obtained (JIP/IEC/2018/076), and all the participants gave written informed consent before the study. Two hundred and sixteen schizophrenia patients (Age-18–50 years), diagnosed based on the Diagnostic and Statistical Manual of Mental disorders-5 criteria, were enrolled in the study. Two hundred and sixteen age, ethnicity, and gender-matched apparently healthy controls were included in the study. Participants with major medical comorbidities were excluded from the study.
Sample size calculation
The sample size was calculated using Genetic Association Study (GAS) Power Calculator (“Home|GAS Power Calculator,” n.d.). The minor allele frequency (MAF) of rs6489630 in South Asian population (MAF = 0.1651) was considered for sample size calculation (“rs6489630 RefSNP Report - dbSNP - NCBI,”). With the disease prevalence of 0.3%,[13] level of significance of 0.05 and power being 0.90 (90%), the sample size was (n) 216 for both cases and control in each group, respectively.
Assessment of disease severity and cognitive functions
The severity of schizophrenia was assessed by the PANSS scale.[14] Addenbrooke cognitive examination-III (ACE-III) scale was used to assess cognitive functions in patients with schizophrenia.[15] In the present study, a cut-off of 82/83 on the ACE-III was used to detect cognitive dysfunction.[16]
Sample collection and biochemical analysis
Five mL of venous blood was collected in an EDTA tube and centrifuged at 3500 rpm for 10 min. Plasma was collected and stored at-40°C and used for the estimation of NT-3 levels. Plasma NT-3 levels were estimated by enzyme-linked immunosorbent assay using the commercially available kit (Elabscience, USA).
Deoxyribonucleic acid (DNA) was extracted from the remaining cell pellet following the protocol provided in the QIAmp DNA Blood mini kit manufactured by Qiagen, Germany.
The working standardized concentration in all the DNA samples was diluted to 50 ng/µ with a 260/280 ratio between 1.8 and 2.0 (quantified using NanoDrop™ 2000). The DNA samples were stored at a temperature of 40°C for further genotyping analysis.
Probes were procured from Thermo Fisher U.S. The sequence of the probe is illustrated in [Supplementary Table 1]. Genotyping of NT-3 gene polymorphism at rs6489630, rs6332 and rs11063714 were analyzed using real-time PCR system BioRad CFX 96, (BioRad Laboratories Inc., USA).
A linkage disequilibrium analysis was performed for the three SNPs of gene which is located on chromosome 12 [Figure 1]. Statistical software Haploview version 4.2 was used to construct the haplotype block based on the confidence interval (CI) approach as shown in [Figure 1]. The block was constructed between the three SNPs of NT-3 gene – rs6332, rs6489630 and rs11063714.
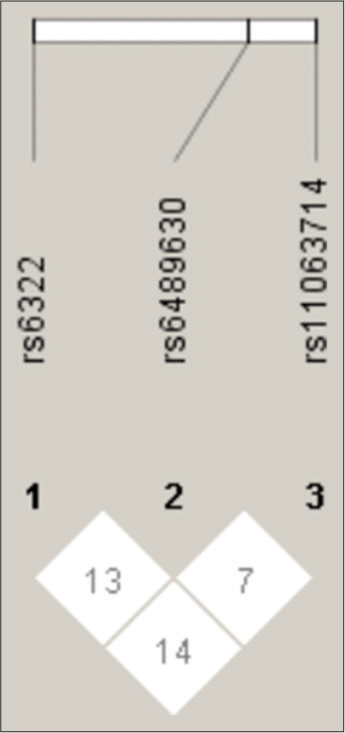
- Linkage Disequilibrium plot of the Neurotrophin-3 gene polymorphisms.
Statistical analysis
The normality of the data was checked by the Kolmogorov– Smirnoff test. The data between cases and controls were compared using Mann–Whitney U test. The relation between NT-3 levels, PANSS, and ACE-III scores was evaluated usingthe Spearman correlation test. The Hardy–Weinberg equilibrium was established using Hardy Weinberg equation (HWE) calculator in the cases and controls. For genotyping analysis, results were expressed in terms of percentage and odds ratio (OR) with a 95% confidence interval and analyzed using Chi-square test using Graph Pad InStat software version 3.0. Genetic model analysis was used to confirm the genetic association. One-way ANOVA and Kruskal–Wallis test were used to compare PANSS, ACE III score, and plasma NT-3 levels between different genotypes and different levels of cognitive scores. Linkage disequilibrium was analyzed using HaploView software version 4.1. A P < 0.05 was considered statistically significant.
RESULTS
[Table 1] shows the general and clinical characteristics like age, gender, BMI, PANSS, ACE-III scores, and NT-3 levels in cases and controls. NT-3 was found to be significantly elevated in cases (P = 0.0001) compared to controls.
| S. No. | Parameter | Control (n=216) | Schizophrenia (n=216) | P |
|---|---|---|---|---|
| 1. | Age (years) | 36.33±8.81 | 35±9.35 | 0.232 |
| 2. | Gender (female/male) | 136/80 | 97/119 | - |
| 3. | Body mass index (kg/m2) | 25.39±3.90 | 25.54±4.14 | 0.697 |
| 4. | Drug naïve/Drug free | - | 119/97 | |
| 5. | Socioeconomic status (upper/lower) | 169/47 | 52/164 | - |
| 6. | NT-3 (ng/L) | 125.25 (94.5–187.72) | 197.50 (133.25–287.0) | 0.0001 |
| 7. | Duration of illness (years) | - | 3 (1–7) | - |
| 8. | Positive symptoms score | - | 15 (10–19.75) | - |
| 9. | Negative symptoms score | - | 16 (11–21) | - |
| 10. | General psychopathological score | - | 28 (21–36) | - |
| 11. | Total PANSS | - | 61.67±22.40 | - |
| 12. | Attention | - | 14 (11–16) | - |
| 13. | Fluency | - | 6 (5–8) | - |
| 14. | Memory | - | 11 (5–18) | - |
| 15. | Language | - | 23 (21–26) | - |
| 16. | Visuospatial abilities | - | 11 (9–14) | - |
| 17. | Total ACE-III | - | 64.63±17.43 | - |
NT-3: Neurotrophin-3, ACE-III: Addenbrooke cognitive examination-III, PANSS: Positive and negative symptom scores
The serum NT-3 levels were significantly correlated with positive symptoms scores (r = −0.23, P = 0.001), general psychopathology scores (r = −0.18, P = 0.007), and total PANSS scores (r = −0.25, P = 0.001). There was no significant association of NT-3 with cognition scores and duration of illness in schizophrenia cases (data available on request).
NT-3 genotypes in schizophrenia and controls
The distribution of NT-3 polymorphism (rs6489630, rs6332, and rs11063714) in schizophrenia cases and controls are shown in [Table 2]. The distribution of rs6489630 and rs6332 was found to be in accordance with HWE in both groups. However, the distribution of rs1163714 was not in agreement with HWE in our study population. Among the genotypes of rs6489630, we observed a significant difference in the frequency of CT genotype between cases and controls and it conferred an increased susceptibility to develop schizophrenia (P = 0.02, OR = 1.631 [1.10–2.43]). The mutant genotype TT also conferred a trend toward the development of schizophrenia, but it was not significant (P = 0.05, OR = 2.5 [1.05–6.19]). Furthermore, minor allele T was found to be associated with increased susceptibility of schizophrenia (P = 0.004, OR = 1.58 [1.16–2.16]). The CT + TT genotype in the dominant model was found to increase the susceptibility (P = 0.006, OR = 1.72 [1.18–2.53]) and CC + TT genotype in the co-dominant model was found to reduce the susceptibility of schizophrenia (P = 0.048, OR = 0.664 [0.450–0.980]). The genotype and allele frequencies of rs6332 and rs11063714 were not significant between the cases and controls.
| Polymorphism | Genotype | Schizophrenia cases (n=216) (%) | Control (n=216), (%) | P | OR (95% CI) |
|---|---|---|---|---|---|
| rs6489630 | CT | 95 (44.0) | 74 (34.3) | 0.02 | 1.631 (1.10–2.43) |
| TT | 16 (7.4) | 8 (3.7) | 0.05 | 2.55 (1.05–6.19) | |
| CC | 105 (48.6) | 134 (62.0) | Ref | 1.58 (1.16–2.16) | |
| Minor allele | T | 127 (29.4) | 90 (20.8) | 0.004 | 1.72 (1.18–2.53) |
| Major allele | C | 305 (70.6) | 342 (79.2) | Ref | 0.48 (0.20–1.15) |
| Dominant model analysis | CT+TT | 111 (51.4) | 82 (37.9) | 0.006 | 0.66 (0.45–0.98) |
| CC | 105 (48.6) | 134 (62.1) | Ref | ||
| Recessive model analysis | CC+CT | 200 (92.6) | 208 (96.3) | 0.140 | |
| TT | 16 (7.4) | 8 (3.7) | Ref | ||
| Codominant model analysis | CC+TT | 121 (56.1) | 142 (65.7) | 0.048 | |
| CT | 95 (43.9) | 74 (33.3) | Ref | ||
| rs6332 | AG | 114 (52.7) | 104 (48.1) | 0.608 | 1.15 (0.75–1.76) |
| AA | 36 (16.8) | 43 (20.0) | 0.743 | 0.87 (0.50–1.52) | |
| GG | 66 (30.5) | 69 (31.9) | Ref | 0.96 (0.74–1.26) | |
| Minor allele | A | 186 (43.1) | 190 (44.0) | 0.836 | 1.67 (0.71–1.6) |
| Major allele | G | 246 (56.9) | 242 (56.0) | Ref | 1.24 (0.76–2.03) |
| Dominant model analysis | AG+AA | 150 (69.4) | 147 (68.0) | 0.835 | 0.83 (0.569–0.121) |
| GG | 66 (30.6) | 69 (32.0) | Ref | ||
| Recessive model analysis | GG+AG | 180 (83.3) | 173 (80.0) | 0.455 | |
| AA | 36 (16.7) | 43 (20.0) | Ref | ||
| Codominant model analysis | GG+AA | 102 (47.2) | 112 (51.8) | 0.386 | |
| AG | 114 (52.8) | 104 (48.2) | Ref | ||
| rs11063714 | AG | 184 (85.5) | 191 (88.5) | 0.393 | 0.7526 (0.42–1.3) |
| GG | 32 (14.8) | 25 (11.5) | Ref | 0.93 (0.71–1.21) | |
| AA | 0 | 0 | - | ||
| Minor allele | A | 184 (42.6) | 191 (44.2) | 0.680 | |
| Major allele | G | 248(57.4) | 241 (55.8) | Ref |
rs6489630: HWE for cases - χ2=0.76, P=0.381; HWE for controls, χ2=0.32, P=0.570. rs6332: HWE for cases - χ2=1.25, P=0.262; HWE for controls, χ2=0.113, P=0.736. rs11063714: HWE for cases - χ2=135.6, P=0.0001; HWE for controls, χ2=118.91, P=0.0001. HWE: Hardy–Weinberg equilibrium, OR: Odds ratio, CI: Confidence interval, NT-3: Neurotrophin-3
When NT-3 levels were compared between different genotypes of rs6489630, rs6332 and rs11063714, we did not observe any significant difference in NT-3 levels between the groups [Supplementary Table 2].
When NT-3 levels were compared between different genotypes of rs6489630, rs6332, and rs11063714, we did not observe any significant difference in NT-3 levels between the groups [Supplementary Table 2].
Among three SNPs, rs6489630 was found to be significantly associated with disease severity (P = 0.004), and rs1063714 was found to be associated with disease severity, but it was not significant (P = 0.08) [Supplementary Table 3].
Among NT-3 genetic variants, rs6332 variant was found to affect cognitive status significantly in schizophrenia (P = 0.040). There was no significant difference in NT-3 levels in schizophrenia patients with normal cognitive function and cognitive impairment [Supplementary Table 4].
Among genotypes of rs6332, memory score was significantly reduced in individuals with AG (P < 0.01) and AA variant (P = 0.03) compared to the GG variant. The total PANSS score, positive symptom score, and general psychopathology score were significantly increased in individuals with the AG variant compared to those with the GG variant (P < 0.05). There was no significant difference in other cognitive domains between the genotypes [Supplementary Table 5].
We did not observe any significant difference in disease severity scores and cognitive scores among the genotypes of rs11063714 and rs6489630.
In the present study, there was no or weak linkage disequilibrium between rs6322 and rs6489630 (D°-0.13), between rs6489630 and rs11063714 (D°-0.07) and between rs6332 and rs11063714 (D°-0.14). A total of eight different haplotype structures were derived from the three SNPs. We did not observe any significant difference in the distribution of haplotypes between the schizophrenia and control subjects (data available on request).
DISCUSSION
In the present study, we found that CT and TT genotypes and minor allele T of rs6489630 variant of the NT-3 gene increase the susceptibility of schizophrenia. The rs6332 variant was found to be associated with cognitive dysfunction in schizophrenia, and memory score was found to be reduced among patients with AG and AA genotypes of rs6332.
Several studies in the past have investigated the association of NT-3 gene polymorphism with schizophrenia, but the results were inconclusive.[12,17] In the present study, we investigated the three SNPs of the NT-3 gene at rs6489630, rs6332, and rs11063714 and their association with the components of cognitive function and disease severity. The genotypic distribution of rs6489630 and rs6332 was found to be in accordance with HWE in both groups suggesting no drift in allelic characteristics from one generation to another. Discordance in the HWE was observed in the genotypic distribution of rs11063714 in both control and schizophrenia casesThe rs6489630 (T > A/C/G) is a functional polymorphism located in an intron on the 3’-side. The rs6332 (G > A/T) is a silent (synonymous) polymorphism located within Pro-NT-3 gene exon 1 and implicated in the regulation of mRNA splicing and protein translation.[18] The rs11063714 (G > A) is a promoter polymorphism located downstream of NT-3 gene which was found to be associated with curve progression in patients with idiopathic scoliosis.[10] Earlier studies have demonstrated the association of rs6489630 and rs6332 with attention deficit hyperactivity disorder and early onset Alzheimer’s disease.[19,20] A previous meta-analyses did not found any association between NT-3 gene polymorphism and schizophrenia.[17] In the present study, we observe that the heterozygous CT variant (P = 0.02), mutant TT variant (P = 0.05), minor variant T (P = 0.004), CT + TT in the dominant model (P = 0.006) of rs6489630 were associated with increased risk of schizophrenia. Furthermore, we found that rs6489630 was significantly associated with the severity of schizophrenia (P = 0.004) and individuals with CC variant were associated with mild illness and CT variant were associated with moderate illness. We did not find any significant difference in genotypes of rs6332 and rs11063714 between schizophrenia and controls, but among genotypes of rs6332, the total PANSS score, positive symptom score and general psychopathology score was significantly increased in individuals with AG variant compared to those with GG variant (P < 0.05). We did not observe any significant difference in NT-3 level between different genotypes of rs6489630, rs6332, and rs11063714 indicating polymorphism of NT-3 gene does not influence plasma NT-3 levels. Linkage disequilibrium analysis showed weak linkage disequilibrium between three SNPs of NT-3 gene and haplotype distribution was not significant between schizophrenia and controls which might be due to the distant location of these loci from one another.
NT-3 levels were enhanced significantly in schizophrenia cases (P < 0.001) and correlated negatively with positive symptoms Score (P = 0.001), general psychopathological Score (P = 0.007), and total PANSS Scores (P = 0.001) suggesting an inverse relationship between NT-3 and severity of schizophrenia. These findings were in accordance with the previous study from our laboratory.[4]
Cognitive dysfunction is an indicator of functional and treatment outcome in schizophrenia and around 70–80% of schizophrenia patients were reported to have cognitive deficits.[21] Aberrant synaptic plasticity was known to be associated with cognitive impairment.[4,5] In the present study, NT-3 levels were neither altered nor associated significantly with components of cognitive dysfunction in schizophrenia which was similar to our previous study.[4] There are paucity of reports in the literature about NT-3 polymorphism and its relation with cognitive impairment in schizophrenia. In the present study, rs6332 variant was found to be significantly associated with cognitive impairment (P = 0.040). Furthermore, memory score was significantly reduced in individuals with AG (P < 0.01) and AA variant (P = 0.03) compared to GG variant. We did not observe any significant association of rs6489630 and rs11063714 with cognitive domains in schizophrenia cases.
The present study has few limitations. The study was conducted only in south Indian population. A larger cohort is needed to validate the results obtained from our study. Only three SNPs of NT-3 gene were studied due to time constraints. Further studies with more SNPs are needed to understand the influence these SNPs in complications of schizophrenia. Cognitive status was not analyzed in controls due to ethical constraints.
CONCLUSION
We conclude that rs6489630 polymorphism of NT-3 gene enhances the risk of developing schizophrenia and was related with severity of the disease. rs6332 polymorphism was associated with cognitive impairment and AG variant of rs6332 was associated with disease severity and memory dysfunction in patients with schizophrenia. The mRNA expression of NT-3 gene and its relation with cognitive dysfunction in schizophrenia needs to be investigated in future studies.
Declaration of patient consent
The authors certify that they have obtained all appropriate consent.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Schizophrenia: A concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30:67-76.
- [CrossRef] [PubMed] [Google Scholar]
- Targeting synaptic plasticity in schizophrenia: Insights from genomic studies. Trends Mol Med. 2021;27:1022-32.
- [CrossRef] [PubMed] [Google Scholar]
- The relationship between cognition and functioning in schizophrenia: A semi-systematic review. Schizophr Res Cogn. 2021;27:100217.
- [CrossRef] [PubMed] [Google Scholar]
- Matrix metalloproteinase-9 increases the risk of cognitive impairment in schizophrenia. Nord J Psychiatry. 2021;75:130-4.
- [CrossRef] [PubMed] [Google Scholar]
- Relationship between neural cell adhesion molecule-1 and cognitive functioning in schizophrenia spectrum disorder. Ind J Clin Biochem. 2020;37:494-8.
- [CrossRef] [PubMed] [Google Scholar]
- Does neurotropin-3 have a therapeutic implication in major depression? Int J Neurosci. 2008;118:1515-22.
- [CrossRef] [PubMed] [Google Scholar]
- Neurotrophic factors in bipolar disorders patients with manic episode. Turk J Med Sci. 2020;50:985-93.
- [CrossRef] [PubMed] [Google Scholar]
- Brain, blood, cerebrospinal fluid, and serum biomarkers in schizophrenia. Psychiatry Res. 2018;265:25-38.
- [CrossRef] [PubMed] [Google Scholar]
- A promoter polymorphism of neurotrophin 3 gene is associated with curve severity and bracing effectiveness in adolescent idiopathic scoliosis. Spine (Phila Pa 1976). 2012;37:127-33.
- [CrossRef] [PubMed] [Google Scholar]
- Polymorphisms of the neurotrophic factor-3 (NTF-3) in Alzheimer's disease: rs6332 associated with onset time and rs6489630 T allele exhibited a protective role. J Neurogenet. 2015;29:183-7.
- [CrossRef] [PubMed] [Google Scholar]
- Novel polymorphisms in the promoter region of the neurotrophin-3 gene and their associations with schizophrenia. Am J Med Genet. 2002;114:304-9.
- [CrossRef] [PubMed] [Google Scholar]
- The burden of mental disorders across the states of India: The Global Burden of Disease Study 1990-2017. Lancet Psychiatry. 2020;7:148-61.
- [Google Scholar]
- Outlier-response pattern checks to improve measurement with the positive and negative syndrome scale (PANSS) Psychiatry Res. 2021;303:114114.
- [CrossRef] [PubMed] [Google Scholar]
- Validation of the Addenbrooke's Cognitive Examination III in frontotemporal dementia and Alzheimer's disease. Dement Geriatr Cogn Disord. 2013;36:242-50.
- [CrossRef] [PubMed] [Google Scholar]
- Validation of the Portuguese version of Addenbrooke's Cognitive Examination III in mild cognitive impairment and dementia. Adv Clin Exp Med. 2018;27:781-6.
- [CrossRef] [PubMed] [Google Scholar]
- Meta-analyses of the association between genetic polymorphisms of neurotrophic factors and schizophrenia. Schizophr Res. 2004;71:353-60.
- [CrossRef] [PubMed] [Google Scholar]
- A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525-8.
- [CrossRef] [PubMed] [Google Scholar]
- Neurotrophin-3 gene, intelligence, and selective attention deficit in a Korean sample with attention-deficit/hyperactivity disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:1065-9.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic association between neurotrophin-3 polymorphisms and Alzheimer's disease in Japanese patients. Dement Geriatr Cogn Dis Extra. 2013;3:272-80.
- [CrossRef] [Google Scholar]
- How should DSM-V criteria for schizophrenia include cognitive impairment? Schizophr Bull. 2007;33:912-20.
- [CrossRef] [PubMed] [Google Scholar]
SUPPLEMENTARY TABLES
| S. No. | rsID | Sequence |
|---|---|---|
| 1. | rs6489630 | TCTATATCAAATTCTCTGCTGTTAG[C/T]TTGGCAAATTAGAATTATGATTTGC |
| 2. | rs6332 | GGCCCGCCAAGTCAGCATTCCAGCC[A/G]GTGATTGCAATGGACACCGAACTGC |
| 3. | rs11063714 | ATAGAGGATAAATTTATTTATGGCC[A/G]ACTCTCAGCCTGTGACCACCCCTCC |
NT-3: Neurotrophin-3
| S. No. | NT-3 levels | Genotypes | P-value | ||
|---|---|---|---|---|---|
| 1. | rs6489630 | CC (n=105) | CT (n=95) | TT (n=16) | |
| NT-3 (pg/mL) | 193 (124–287) | 205 (153–328) | 181 (152–248) | 0.371 | |
| 2. | rs6332 | GG (n=66) | AG (n=114) | AA (n=36) | |
| NT-3 (pg/mL) | 198 (126.50–256.75) | 207 (31.50–3031.75) | 169 (124.50–252.50) | 0.338 | |
| 3. | rs11063714 | AG (n=184) | GG (n=32) | - | |
| NT-3 (pg/mL) | 200 (143–286.50) | 161 (79.75–355.00) | 0.338 |
NT-3: Neurotrophin-3
| Genotype | Schizophrenia with mild illness (PANSS: 30–58) (n=107) (%) |
Schizophrenia with moderate illness (PANSS: 59–74) (n=54) (%) |
Schizophrenia with marked illness (PANSS: 75-94) (n=37) (%) |
Schizophrenia with severe illness (PANSS: >95) (n=18) (%) |
P-value |
|---|---|---|---|---|---|
| rs6489630 | |||||
| CC | 60 (56) | 22 (40.7) | 17 (46) | 6 (33.3) | 0.004 |
| CT | 37 (34.5) | 32 (59.3) | 18 (48.6) | 8 (44.4) | |
| TT | 10 (9.5) | 0 | 2 (5.4) | 4 (22.3) | |
| rs6332 | |||||
| GG | 38 (35.5) | 17 (32) | 6 (16) | 5 (27.7) | 0.452 |
| AG | 51 (47.7) | 28 (52) | 24 (65) | 11 (61.1) | |
| AA | 18 (16.8) | 9 (16) | 7 (19) | 2 (11.2) | |
| rs1063714 | |||||
| AG | 95 (88.8) | 48 (89) | 28 (75.7) | 13 (72.3) | 0.08 |
| GG | 12 (11.2) | 6 (11) | 9 (24.3) | 5 (27.7) | |
NT-3: Neurotrophin-3, PANSS: Positive and negative symptom scores
| Genotype | Schizophrenia cases | P-value | ||
|---|---|---|---|---|
| Severe cognitive impairment (ACE III score: 0–61) (n=88) (%) | Mild cognitive impairment (ACE III score: 62–81) (n=93) (%) | Normal cognitive function (ACE III score: 82–100) (n=35) (%) | ||
| rs6489630 | ||||
| CC | 40 (45.5) | 50 (53.7) | 15 (42.8) | 0.377 |
| CT | 42 (47.7) | 38 (40.8) | 15 (42.8) | |
| TT | 6 (11.8) | 5 (5.5) | 5 (14.2) | |
| rs6332 | ||||
| GG | 20 (22.7) | 28 (30.1) | 18 (51.4) | 0.040 |
| AG | 51 (57.9) | 49 (52.7) | 14 (40.) | |
| AA | 17 (19.3) | 16 (17.2) | 3 (8.6) | |
| rs11063714 | ||||
| AG | 73 (82.9) | 78 (83.8) | 32 (91.4) | 0.250 |
| GG | 15 (17.1) | 15 (16.2) | 3 (9.6) | |
| NT-3 (ng/L) | 187.0 (117.0–289.0) | 134.0 (206.0–276.0) | 211.0 (137.0–348.0) | 0.443 |
NT-3: Neurotrophin-3, ACE-III: Addenbrooke cognitive examination-III
| S. No. | GG (n=66) | AG (n=114) | AA (n=36) | |
|---|---|---|---|---|
| 1. | Attention | 14±3.93 | 12.92±4.16 | 13.11±3.28 |
| 2. | Memory | 14 (7.7–23.2) | 9.5* (4–16) | 7a(4–16) |
| 3. | Fluency | 6.8±2.7 | 6.0±2.8 | 6.0±2.49 |
| 4. | Language | 22.59±4.97 | 22.18±4.1 | 22.19±3.17 |
| 5. | Visuospatial abilities | 11.38±3.29 | 11.54±3.37 | 11.97±2.75 |
| 6. | Total ACE-III score | 68.48±19.33 | 62.98±16.82 | 62.78±14.74 |
| 7. | Positive symptom score | 13.8±6.4 | 16.65±6.62** | 15.0±6.5 |
| 8. | Negative symptom score | 15.83±7.14 | 17.14±7.6 | 15.08±6.31 |
| 9. | General psychopathological score | 27.61±10.17 | 31.66±12.02** | 28.33±9.85 |
| 10. | Total PANSS | 57.24±20.89 | 65.45±23.0** | 54.42±21.63 |






