Translate this page into:
Machine learning in action: Revolutionizing intracranial hematoma detection and patient transport decision-making
*Corresponding author: Ahmed Al Menabbawy, Department of Neurosurgery, Cairo University, Cairo, Egypt. Ahmed.almenabbawy@med.uni-greifswald.de
-
Received: ,
Accepted: ,
How to cite this article: El Refaee E, Ali TM, Al Menabbawy A, Elfiky M, El Fiki A, Mashhour S, et al. Machine learning in action: Revolutionizing intracranial hematoma detection and patient transport decision-making. J Neurosci Rural Pract. 2024;15:62-8. doi: 10.25259/JNRP_93_2023
Abstract
Objectives:
Traumatic intracranial hematomas represent a critical clinical situation where early detection and management are of utmost importance. Machine learning has been recently used in the detection of neuroradiological findings. Hence, it can be used in the detection of intracranial hematomas and furtherly initiate a management cascade of patient transfer, diagnostics, admission, and emergency intervention. We aim, here, to develop a diagnostic tool based on artificial intelligence to detect hematomas instantaneously, and automatically start a cascade of actions that support the management protocol depending on the early diagnosis.
Materials and Methods:
A plot was designed as a staged model: The first stage of initiating and training the machine with the provisional evaluation of its accuracy and the second stage of supervised use in a tertiary care hospital and a third stage of its generalization in primary and secondary care hospitals. Two datasets were used: CQ500, a public dataset, and our dataset collected retrospectively from our tertiary hospital.
Results:
A mean dice score of 0.83 was achieved on the validation set of CQ500. Moreover, the detection of intracranial hemorrhage was successful in 94% of cases for the CQ500 test set and 93% for our local institute cases. Poor detection was present in only 6–7% of the total test set. Moderate false-positive results were encountered in 18% and major false positives reached 5% for the total test set.
Conclusion:
The proposed approach for the early detection of acute intracranial hematomas provides a reliable outset for generating an automatically initiated management cascade in high-flow hospitals.
Keywords
Artificial intelligence
Deep learning
Automatic intracranial hematoma detection
Patient selection
Patient referral system
INTRODUCTION
Timely management of intracerebral hematomas is very crucial in preventing secondary brain injury and the outcome. Application of an effective neurotrauma management system is a cardinal need in high-flow areas that warrant intensely demanding assignments.[1-3] During the conception of a primed management protocol, the formulation of low-cost tools for better communication and faster management is fundamental, together with real-time registries of the number of patients, surgeries, and medical personnel.[4,5] Machine learning (ML) has been initially introduced and firstly accepted in the neurosurgical community. It mainly supports various objectives starting with the quantitative measurement of the radiological findings, reaching the outcome analysis.[6] Intracranial hematomas warrant critical clinical situations where early detection and management would be utterly essential.[7,8] In a wide range of health-care facilities in middle- and low-income countries, the emergency management protocols need enhancements so that they can be routed toward a fast-track mainframe. Due to the retrenchment of a fully integrated information system, it might be needed to develop instantaneous detection tools that would initiate a management cascade of patient transfer, diagnostics, admission, and emergency intervention that might be in some instances lifesaving. This study aims to develop a new diagnostic tool depending on artificial intelligence (AI), where the machine can learn how to detect the hematomas instantaneously, and furtherly automatically start a cascade of actions that support the management protocol depending on the early diagnosis.
MATERIALS AND METHODS
A team of medical researchers in the fields of Neurosurgery, Trauma, Radiology, and Biomedical Engineering designed the plot as a staged model to develop the early diagnostic model with direct clinical integration in three stages: The first stage of initiating and training the machine with provisional detection of its accuracy; the second stage is supervised use of this model at our local institute with more detection of fallacies and correcting them; and the third stage of using this model in other primary or secondary care referring hospitals where a computerized tomography (CT) is available with no neurosurgical team present ([Figure 1] which illustrates the management cascade of head trauma).
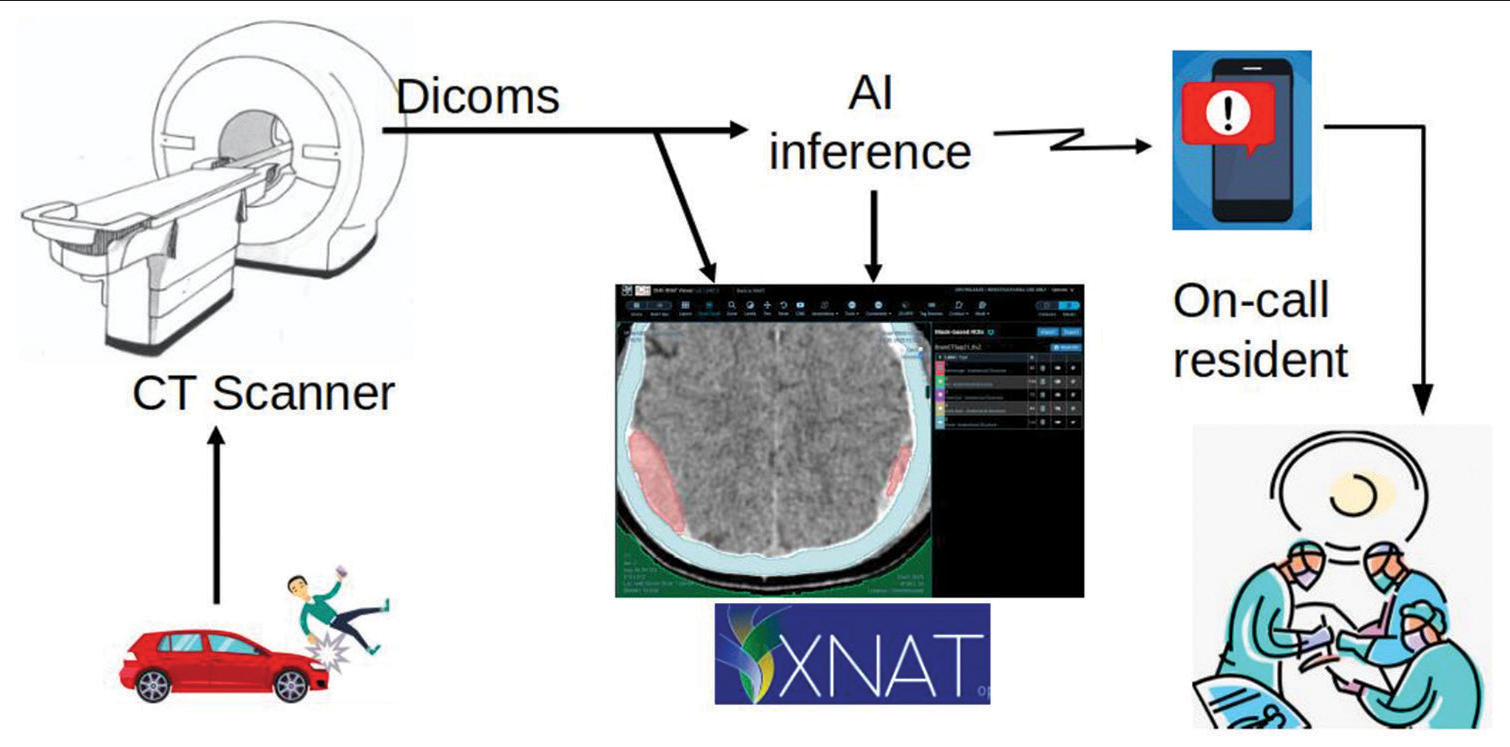
- Management cascade of head trauma. Victims of head trauma are transferred to a hospital where a computerized tomography scan is done. Being connected to the automated intelligence machine, intracranial hematomas are instantaneously detected with immediate notification of the oncall resident. This allows early surgery or transfer to the nearest neurosurgery center.
Our system is designed to run with minimal cost in the emergency department for head trauma cases. Once a head CT is performed, digital imaging and communications in medicine (DICOM) images are pushed to the XNAT system and AI inference. XNAT is a research PACS system.[9,10] We used Clara Train AIAA inference server to run the AI models. The segmentation result is, then, pushed to XNAT for review. During the next phase, once intracranial hemorrhage is detected, an automatically generated message would be sent instantaneously to the neuroemergency team.
To get ahead, we started using the CQ500 public dataset. However, unlike their initial study, which created a neural network model to classify 2D slices,[11] we developed a 3D segmentation model here. Clara Train SDK was used to train a SegResNet model.[12] SegResNet architecture is an encoder-decoder network. The encoding part increases the number of filters while decreasing the field of view, while the decoding uses upsampling to increase the resolution of the original input-sized image. Each block of the encoding and decoding blocks is a residual block. Output of the encoding blocks is appended to the decoding blocks creating a bypass. Multiple augmentation transformations were used from Clara Train SDK as resampling, cropping, rotation, and intensity variation. Weighted dice loss was used during training to accommodate for pixel class imbalance.
Study design
We used two datasets in this project: The CQ500 dataset which is publicly available, and the dataset retrospectively collected from the emergency department of our local institute. Our study got an ethical approval (N-123-2020) from the Faculty Local Ethical Committee. A consolidation of public dataset and randomized testing imaging samples from the institutional dataset was agreed to be ample for stage one. Images from our institution were obtained from the emergency department under the supervision of the Radiology, Neurosurgery, and Trauma surgery teams. The data were collected and experimented between December 2020 and October 2021.
The CQ500 dataset contained almost 500 brain CTs with different diagnoses including brain fracture, hemorrhage, and subdural hematoma. A senior radiologist went through the dataset and identified 80 cases of hemorrhage. Only 58 out of the 80 were annotated; therefore, the CQ500 dataset was split into three sets: 38 for training 20 for validation, and the remaining 22 cases were kept for testing. We also identified 11 cases without any hemorrhage to act as normal controls which will help identify the false positives alerts to the system. This increased the testing dataset to 33 cases.
The local institute dataset contained 42 patients acquired from a GE Medical System machine with the following specifications: FOV 512 × 512; peak kilovoltage of 140; exposure time of 791 ms; exposure of 5 mAs; X-ray tube current of 165 mA; focal spots of 0.7 mm; slice thickness ranging from 0.5 mm to 1.5 mm with space between slices from 0 to 7.5mm; and in-plan spacing of 0.5 mm to 1 mm. All patients from our local institute were collected from our neurosurgical emergency database. However, post-operative cases, those with chronic subdural hematomas, or cases with other pathologies (e.g., tumors) were excluded, reaching a number of 27 cases valid for testing.
Annotation
For annotation, we used the XNAT research PACS system.[9,10] XNAT was configured to convert DICOM images to Nifti format which would be used for training. The XNAT-open health imaging foundation (OHIF) plugin allowed clinicians to segment different regions of the brain. For each patient, multiple structures were annotated: Air outside the skull, skull bone, brain ventricles, and the hemorrhage. Since manual annotation is tedious and time-consuming, the air, bone, and ventricle were segmented by a trained non-clinician with 15 years of experience in radiological imaging. Hemorrhage was segmented by a neurosurgeon with 10 years of experience. XNAT-OHIF saved the annotation as DICOM-SEG objects which were automatically converted to nifit masks using XNAT.
Evaluation
To measure the accuracy of AI segmentation and ground truth annotation during training, the dice coefficient was used. The dice coefficient is defined as twice the intersection of the ground truth and the AI segmentation divided by the sum of the AI segmentation region and the ground truth region. For clinical evaluation, the XNAT system was used, the XNAT-OHIF viewer plugin was used to view the images while the rapid reader plugin was used to evaluate only the hemorrhage lesions. For each patient, the AI accuracy of hemorrhage lesions was evaluated on two dimensions: AI detection accuracy and false-positive errors according to [Table 1] which illustrates different grades of evaluation. Hemorrhage lesions were assessed by two neurosurgeons with 10- and 18-year experience, respectively. Any conflict was solved through consensus between the medical research team.
| Evaluation of lesion detection | Detection percentage | False-positive evaluation | Number of FP regions |
|---|---|---|---|
| Excellent | more than 80 | No false-positive detected | 0% FP |
| Good | 50–80 | Acceptable | 1 small region |
| Fair | 20–50 | Moderate | 2–3 small regions |
| Poor | <20 | Major | >3 or large regions |
FP: False positive.
Experiments
Multiple Segrest models were trained using initial filter sizes of 32, 16, and 8. We tried different resampling resolutions of 0.5 × 0.5 × 1 mm3, 1 × 1 × 1 mm3, as well as 1 × 1 × 2mm3. We also tried different crop sizes of 128 × 128 × 32, 256 × 256 × 64, and 384 × 384 × 64 as well as different increment factors of 2 (standard practice) and 1.5. SegResNet model had an initial 16 filters and an increase factor of 1.5. Brain volumes were resampled to 0.5 × 0.5 × 1 mm3, and intensity clipping transformation was used to clip intestines outside the Hunsfild units of (−100.20) while mapping it to (0.1) intensities range. Training was performed on crops of 384 × 384 × 64 pixels around foreground pixels. The following augmentation transformations were applied: Random flips around the sagittal directions with probability of 0.5; random rotation of −50° to +50° with probability of 0.6 in the axial direction; and random shift intensity with an offset of 0.1 with probability of 0.5. Adam optimizer was used with a learning rate of 0.0001, batch size of 4. Loss function was weighted dice loss with weights of 0.03 for background, 0.61 for hemorrhage, 0.06 for air, 0.15 for ventricles, and 0.15 for bone. 300 epochs of training with validation every 20 epochs were completed in 4 h on a single V100 32 GB GPU. The best model was selected based on the dice score of the hemorrhage label.
RESULTS
Models trained with small initial filters as 8 or small crop size of 128 × 128 × 32 compromised accuracy. Using a larger initial filter size of 16, 32 needed more memory than the 6 GB available in our laptop system. The model with the initial filter size of 16 with a smaller increment factor of 1.5 and crop size of 384 × 384 × 64 pixels resulted in acceptable results while keeping the model size small enough to fit into the 6 GB GPU memory. All the following results are specific to this model. The mean dice score was 0.83 evaluated over the validation dataset of CQ500. Dice per label was 0.95 for background, 0.56 for hemorrhage, 0.98 for air, 0.76 for ventricles, and 0.93 for the bone. All images in the results section show AI prediction of hemorrhage in red, air in green, bone in blue, and ventricles in magenta.
Clinical evaluation for the CQ500 test set and local dataset is shown in Table 2. In the CQ500 test set, intracranial hemorrhage was detected in 31 of 33 cases (94%) with 76% excellent and 18% good detection. Poor detection was present in only 2 cases (6%). Regarding positive results, the results were acceptable in 88% of the cases with moderate false positives reaching 12% (4/33) and no single major false-positive result.
| Detection | CQ500 | Local dataset | Total | False positive | CQ500 | Local dataset | Total |
|---|---|---|---|---|---|---|---|
| Count (%) | Count (%) | Count (%) | Count (%) | Count (%) | Count (%) | ||
| Excellent | 25 (76) | 19 (70) | 44 (73) | No false positive detected | 16 (48) | 10 (37) | 26 (43) |
| Good | 6 (18) | 4 (15) | 10 (16) | Acceptable | 13 (39) | 7 (26) | 20 (33) |
| Fair | 0 (0) | 2 (7) | 2 (3) | Moderate | 4 (12) | 7 (26) | 11 (18) |
| Poor | 2 (6) | 2 (7) | 4 (6) | Major | 0 (0) | 3 (11) | 3 (5) |
Left shows detection accuracy while the right shows false-positive detection evaluation
In our local institute dataset, detection was present in 25 of 27 cases (93%) with 70% excellent, 15% good, and 7% fair detection. Poor detection was present in 2 cases (7%). Positive results were satisfactory and acceptable in 63% (17/27) while moderate false positives reached 12% (7/27) and major false-positive results of 11% (3/37).
Our AI model has adequately recognized acute intracranial hematomas with different sizes [Figure 2], ranging from small punctate lobar hemorrhage [Figure 2a], large-sized hemorrhage [Figure 2b], and medium-sized ones [Figure 2c]. Furthermore, it managed in detection of both supratentorial [Figure 2a, c and d] and infratentorial hemorrhage [Figure 2b]. Figure 3 shows the complete detection of different types of hematomas (Extradural – subdural – intraparenchymal – intraventricular) from our local hospital data.
Although the detection was not optimal in some cases [Figure 4], the hematoma was partially detected and would be valid for further steps.
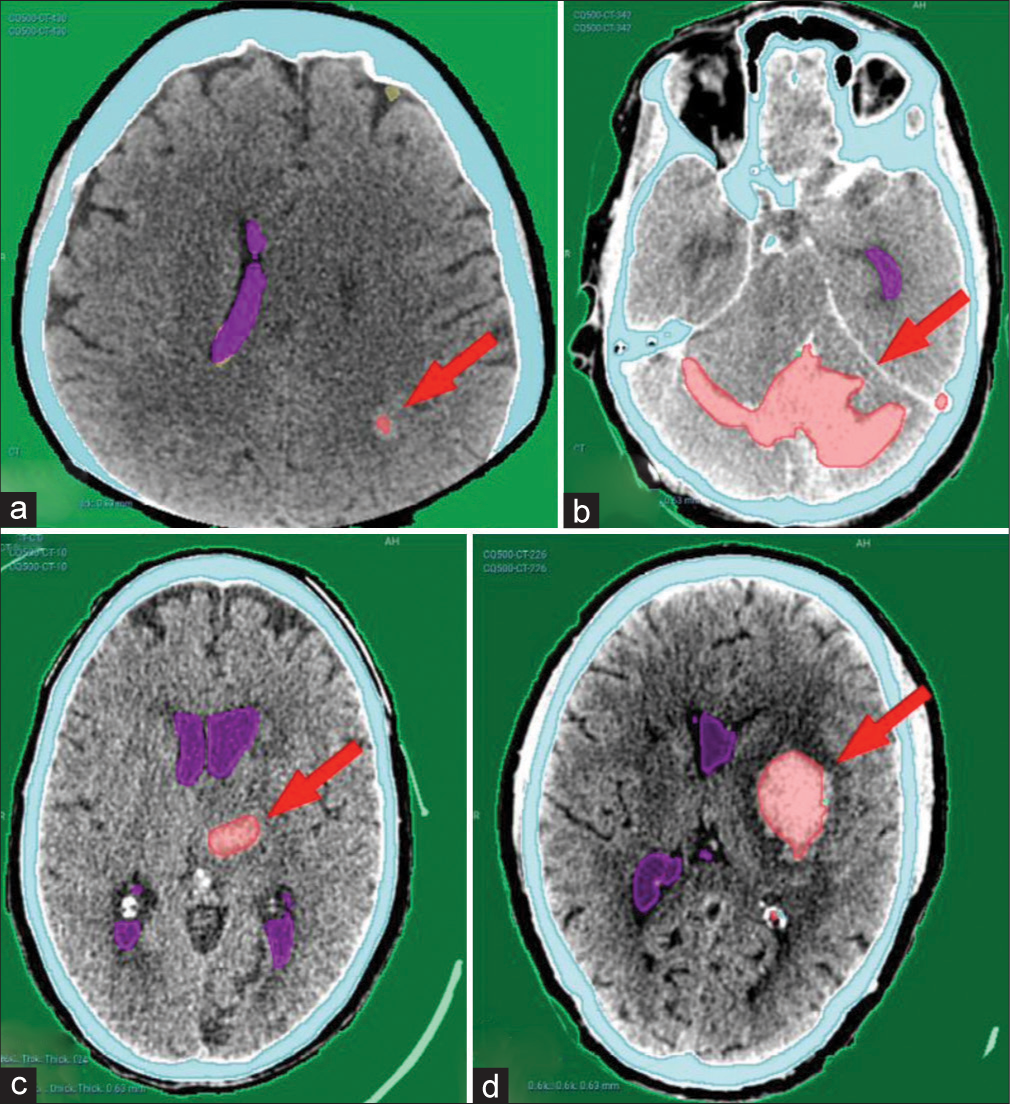
- Examples from the CQ500 test dataset showing complete detection of different hematoma locations and sizes. (a) Left parieto-occipital punctate hematoma. (b) Bilateral cerebellar large hematoma. (c) Left thalamic hematoma. (d) Left basal ganglia hematoma. Red arrows pointing to the hematomas.
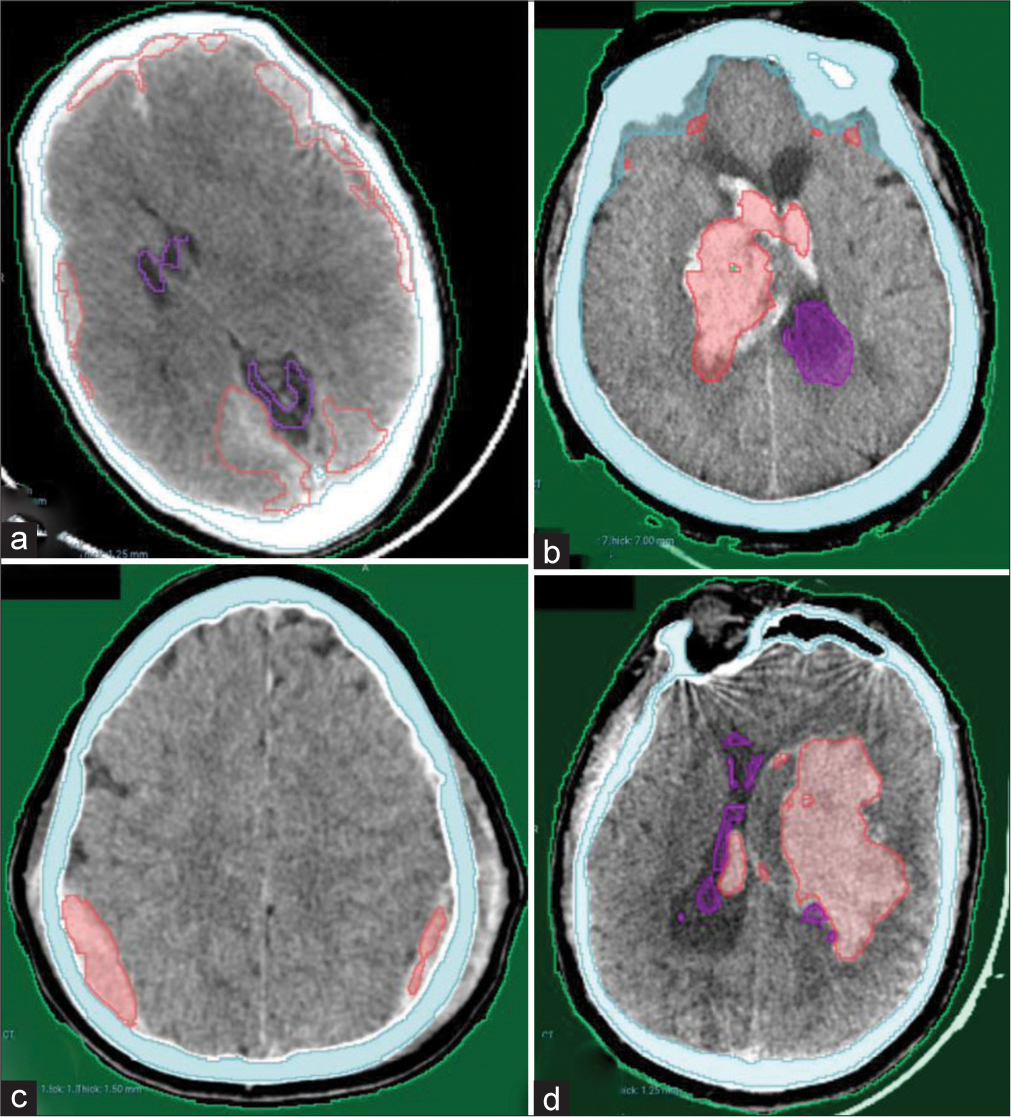
- Examples from our local hospital test dataset showing complete detection of different hematoma locations and sizes. (a) Bilateral acute subdural hematomas (b) Right thalamic hematoma with intraventricular extension. (c) Bilateral parietal extradural hematomas. (d) Left large deep intracerebral hematoma with small intraventricular extension.
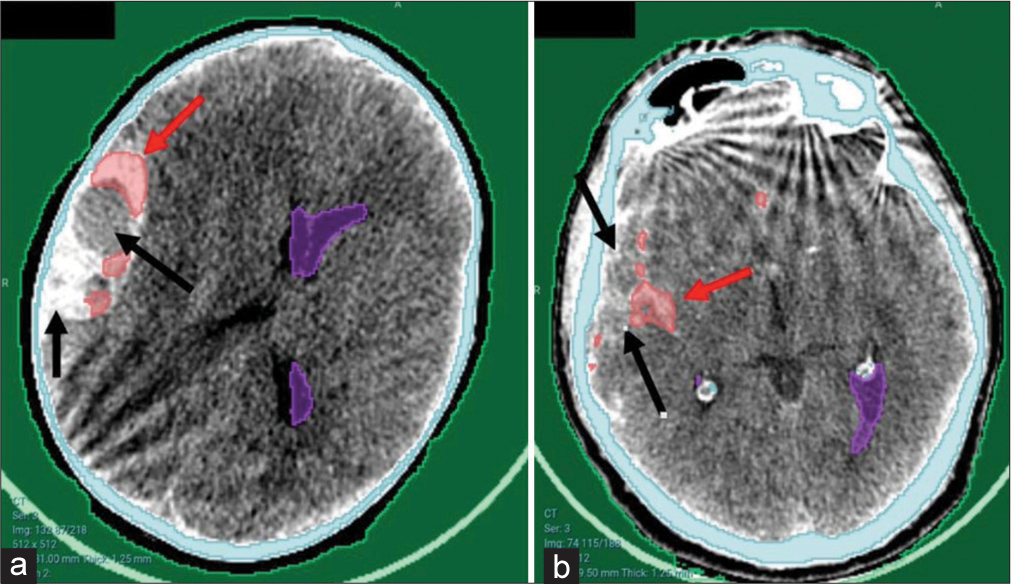
- Examples of partial detection of hematomas from the local hospital. (a) Large right temporoparietal hyperacute extradural hematoma. (b) Subarachnoid hemorrhage in the right Sylvian fissure. Note the bony artifacts present. Arrows poiting to hematoma sites. Red arrows referring to successfully detected areas of the hematoma while the black arrows refer to the missed areas.
However, the model faced difficulties detecting older hematomas, as shown in Figure 5, where a calcified left frontal extradural hematoma [Figure 5a] or a subacute right frontal and occipital subdural hematoma [Figure 5b] were missed.
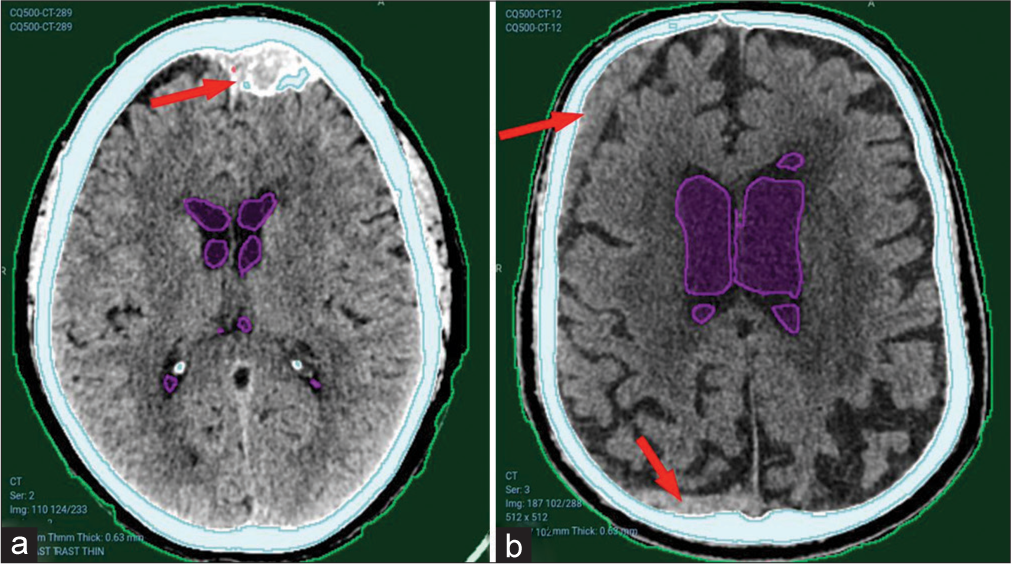
- Undetected hematomas from CQ500 dataset. (a) Left frontal old calcified extradural hematoma. Red arrows pointing to the hematomas. (b) Right frontal and occipital subacute subdural hematomas. Red arrows pointing to bleeding sites that were not detected.
False-positive results [Figure 6] were mostly in dural folds including the tentorium, falx cerebri, or in cerebral venous sinuses, as shown in Figure 6a and b. Other minor false-positive results were found in a minority of cases, as shown in Figure 6c.
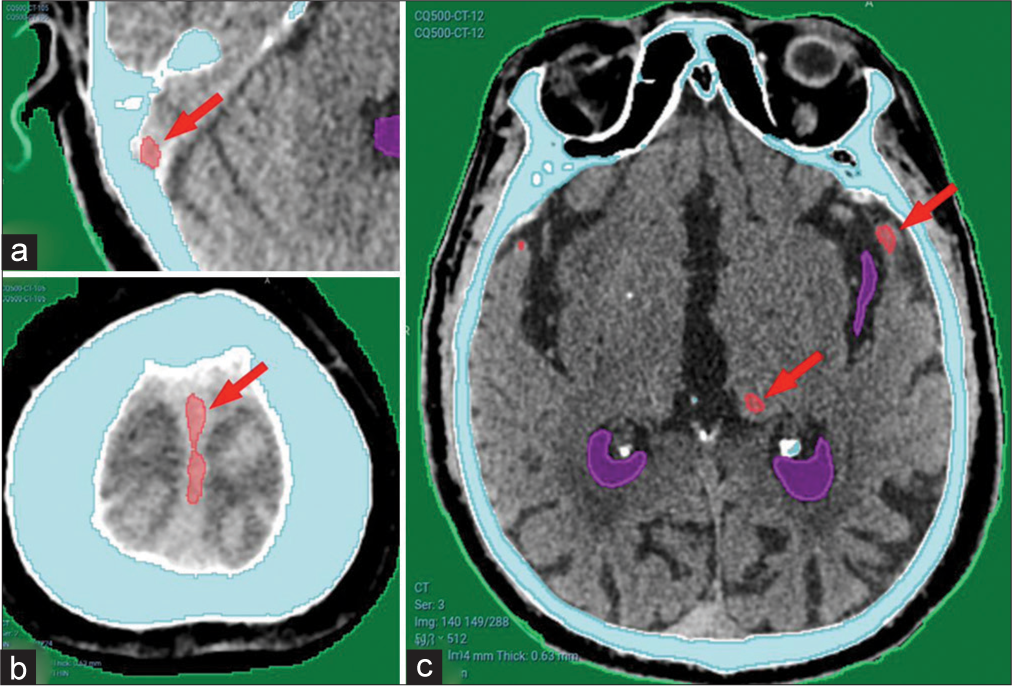
- (a) Showing minor false-positive results from CQ500 dataset in the sigmoid sinus, (b) falx cerebri, and (c) basal ganglia calcification. Red arrows pointing to areas falsely detected as hematomas.
DISCUSSION
Despite the heterogeneity of the CT findings in neurotrauma, the introduction of ML might be pivotal in certain situations as it saves precious time from hematoma detection to intervention.[13] Several studies inspected the competence of the machine to measure the volume of the intracranial hemorrhage,[14,15] together with special concerns to demarcate the brain edema after intracranial insults.[13,16,17] However, our concern was directed toward implementing the ML tools to settle the current challenges and therefore, early detection of intracranial hemorrhage was the first target. Integration of machine detection in clinical workflow would accelerate decision-making; however, accurate results would be crucial for competent clinical arrangements.[18] Instantaneous machine detection of intracranial hemorrhage can initiate an important cascade that serves in the early and effective management of critical cases.
This initiative started with supervised deep learning steps to prepare the machine to detect the early intracranial hematomas depending on the public data set and samples from our local university hospital (being a tertiary referral center) as a first stage. For accurate evaluation of the accuracy of various pathologies, a further step on local cases is warranted. We believe that this should proceed in 3 stages: The first stage of initiating and training the machine with the provisional evaluation of its accuracy (which we have already done), the second stage of supervised use of this model in our local university hospital (human check to validate the machine detection) with more detection of fallacies and correcting them, and a third stage of using this model in other primary or secondary care referring hospitals where a CT is available with no neurosurgical team present. Afterward creation of a network between the CT machines in tertiary care hospitals and referring hospitals from one side, and the neurosurgical team from the other side is planned. This network will lead to early detection and notification to the neurosurgical team in the tertiary hospital; hence, early transfer and management can be initiated which is crucial in head trauma cases.
In an analysis of the current stage and results, a valid outcome was encountered in most cases as previously mentioned before, where the machine detected the presence of an intracranial hematoma in most (93–94%) of the cases tested with a variable range of the volumetric measurements that were detected which is quite satisfactory.[4] It is worth noting that the false-positive results were encountered mostly with dural folds (tentorium and falx cerebri), cerebral venous sinuses (transverse and sigmoid sinuses), and basal ganglia calcifications. False-positive results were also more encountered in our local healthcare facility CT scans than the public dataset due to the lower image quality and the bony artifacts present. False-negative results were encountered only in older hematomas including subacute, chronic, or calcified hematomas. These false-negative results are non-significant when putting into consideration that we are primarily targeting recent head trauma cases (that may develop acute hematomas).
Therefore, the current results are considered supportive to proceed to the second stage of supervised use of this model in our local university hospital. In this second stage, all head trauma cases undergoing CT brain will be used, false-positive and -negative results can be corrected by the available neurosurgical team and improvement of the training process can be performed. This will lead to the enhancement of the accuracy of this model and will, then, encourage going on with the third stage of its generalization to primary and secondary healthcare centers. We believe that using this model would be of maximum benefit in low-middle-income countries where many health-care centers dealing with trauma lack a neurosurgical team.
Study limitations
In the frame of the current accomplishments during the initial phase, the sample size was constrained to assess the chance of the current procedure to touch a prospective that would allow it to step toward further phases. The total number of patient images was split to train, validate, and test the model. Normal controls were only available from the public dataset however with favorable outcomes. Therefore, the current results are worthwhile in comparison to the limited sample size. In all the tested samples, no single false-negative result was encountered in acute head trauma cases which validate the current investigation to be transferred to stage two without a risk to miss cases with acute intracranial hematomas and no major false positives. The main limitation is the relatively high number of minor false-positive results that would improve with more training of the model in the near future.
CONCLUSION
The proposed approach for integrating the machine in the early detection of acute intracranial hematomas provides a reliable outset for generating an automatically-initiated management cascade in high-flow hospitals.
Ethical approval
The study is approved by the Faculty Local Ethical Committee, number (N-123-2020).
Declaration of patient consent
Patient’s consent not required as patients identity is not disclosed or compromised.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Secondary gains: Advances in neurotrauma management. Emerg Med Clin North Am. 2018;36:107-33.
- [CrossRef] [PubMed] [Google Scholar]
- Guidelines for the management of severe head injury. Part 1. Neurotrauma system and neuroimaging. Zh Vopr Neirokhir Im N N Burdenko. 2015;79:100-6.
- [CrossRef] [PubMed] [Google Scholar]
- Development of guidelines for the management of severe head injury. J Neurotrauma. 1995;12:907-12.
- [CrossRef] [PubMed] [Google Scholar]
- Strengthening neurotrauma care systems in low and middle income countries. Brain Inj. 2013;27:262-72.
- [CrossRef] [PubMed] [Google Scholar]
- Essential neurosurgical workforce needed to address neurotrauma in low-and middle-income countries. World Neurosurg. 2019;123:295-9.
- [CrossRef] [PubMed] [Google Scholar]
- Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318-28.
- [CrossRef] [PubMed] [Google Scholar]
- Big data, artificial intelligence, and machine learning in neurotrauma In: Leveraging biomedical and healthcare data. Netherlands: Elsevier; 2019. p. :53-75.
- [CrossRef] [Google Scholar]
- Unsupervised machine learning reveals novel traumatic brain injury patient phenotypes with distinct acute injury profiles and long-term outcomes. J Neurotrauma. 2020;37:1431-44.
- [CrossRef] [PubMed] [Google Scholar]
- Integrating the OHIF viewer into XNAT: Achievements, challenges and prospects for quantitative imaging studies. Tomography. 2022;8:497-512.
- [CrossRef] [PubMed] [Google Scholar]
- Integration of XNAT/PACS, DICOM, and research software for automated multi-modal image analysis. Proc SPIE Int Soc Opt Eng. 2013;8674
- [CrossRef] [PubMed] [Google Scholar]
- Deep learning algorithms for detection of critical findings in head CT scans: A retrospective study. Lancet. 2018;392:2388-96.
- [CrossRef] [PubMed] [Google Scholar]
- Available from: https://catalog.ngc.nvidia.com/orgs/nvidia/containers/clara-train-sdk [Last accessed on 2021 Dec 30]
- Automatic quantification of computed tomography features in acute traumatic brain injury. J Neurotrauma. 2019;36:1794-803.
- [CrossRef] [PubMed] [Google Scholar]
- Fully automated segmentation algorithm for hematoma volumetric analysis in spontaneous intracerebral hemorrhage. Stroke. 2019;50:3416-23.
- [CrossRef] [PubMed] [Google Scholar]
- A robust deep learning segmentation method for hematoma volumetric detection in intracerebral hemorrhage. Stroke. 2022;53:167-76.
- [CrossRef] [PubMed] [Google Scholar]
- Automated quantification of cerebral edema following hemispheric infarction: Application of a machine-learning algorithm to evaluate CSF shifts on serial head CTs. Neuroimage Clin. 2016;12:673-80.
- [CrossRef] [PubMed] [Google Scholar]
- Analysis of medical images using machine learning techniques In: Graph learning and network science for natural language processing. United States: CRC Press; 2022. p. :231-54.
- [CrossRef] [Google Scholar]
- Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit Med. 2018;1:9.
- [CrossRef] [PubMed] [Google Scholar]







